tadA, an essential tRNA-specific adenosine deaminase from Escherichia coli
- PMID: 12110595
- PMCID: PMC126108
- DOI: 10.1093/emboj/cdf362
tadA, an essential tRNA-specific adenosine deaminase from Escherichia coli
Abstract
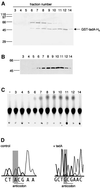
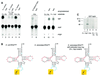
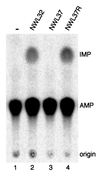
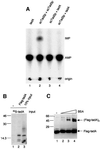
We report the characterization of tadA, the first prokaryotic RNA editing enzyme to be identified. Escherichia coli tadA displays sequence similarity to the yeast tRNA deaminase subunit Tad2p. Recombinant tadA protein forms homodimers and is sufficient for site-specific inosine formation at the wobble position (position 34) of tRNA(Arg2), the only tRNA having this modification in prokaryotes. With the exception of yeast tRNA(Arg), no other eukaryotic tRNA substrates were found to be modified by tadA. How ever, an artificial yeast tRNA(Asp), which carries the anticodon loop of yeast tRNA(Arg), is bound and modified by tadA. Moreover, a tRNA(Arg2) minisubstrate containing the anticodon stem and loop is sufficient for specific deamination by tadA. We show that nucleotides at positions 33-36 are sufficient for inosine formation in mutant Arg2 minisubstrates. The anticodon is thus a major determinant for tadA substrate specificity. Finally, we show that tadA is an essential gene in E.coli, underscoring the critical function of inosine at the wobble position in prokaryotes.
Figures

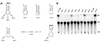
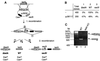
References
-
- Altman S. and Kirsebom,L. (1999) Ribonuclease P. In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
-
- Auxilien S., Crain,P.F., Trewyn,R.W. and Grosjean,H. (1996) Mechanism, specificity and general properties of the yeast enzyme catalysing the formation of inosine 34 in the anticodon of transfer RNA. J. Mol. Biol., 262, 437–458. - PubMed
-
- Bass B.L. (1997) RNA editing and hypermutation by adenosine deamination. Trends Biochem. Sci., 22, 157–162. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

