Crystal structure of human 53BP1 BRCT domains bound to p53 tumour suppressor
- PMID: 12110597
- PMCID: PMC126127
- DOI: 10.1093/emboj/cdf383
Crystal structure of human 53BP1 BRCT domains bound to p53 tumour suppressor
Erratum in
- EMBO J 2002 Nov 1;21(21):5953
Abstract
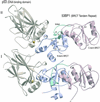
The BRCT (BRCA1 C-terminus) is an evolutionary conserved protein-protein interacting module found as single, tandem or multiple repeats in a diverse range of proteins known to play roles in the DNA-damage response. The BRCT domains of 53BP1 bind to the tumour suppressor p53. To investigate the nature of this interaction, we have determined the crystal structure of the 53BP1 BRCT tandem repeat in complex with the DNA-binding domain of p53. The structure of the 53BP1-p53 complex shows that the BRCT tandem repeats pack together through a conserved interface that also involves the inter-domain linker. A comparison of the structure of the BRCT region of 53BP1 with the BRCA1 BRCT tandem repeat reveals that the interdomain interface and linker regions are remarkably well conserved. 53BP1 binds to p53 through contacts with the N-terminal BRCT repeat and the inter-BRCT linker. The p53 residues involved in this binding are mutated in cancer and are also important for DNA binding. We propose that BRCT domains bind to cellular target proteins through a conserved structural element termed the 'BRCT recognition motif'.
Figures





References
-
- Bargonetti J., Manfredi,J.J., Chen,X., Marshak,D.R. and Prives,C. (1993) A proteolytic fragment from the central region of p53 has marked sequence-specific DNA-binding activity when generated from wild-type but not from oncogenic mutant p53 protein. Genes Dev., 7, 2565–2574. - PubMed
-
- Bork P., Hofmann,K., Bucher,P., Neuwald,A.F., Altschul,S.F. and Koonin,E.V. (1997) A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J., 11, 68–76. - PubMed
-
- Brünger A.T. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr., 54, 905–921. - PubMed
-
- Callebaut I. and Mornon,J.P. (1997) From BRCA1 to RAP1: a widespread BRCT module closely associated with DNA repair. FEBS Lett., 400, 25–30. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

