Comprehensive detection of genomic duplications and deletions in the DMD gene, by use of multiplex amplifiable probe hybridization
- PMID: 12111668
- PMCID: PMC379168
- DOI: 10.1086/341942
Comprehensive detection of genomic duplications and deletions in the DMD gene, by use of multiplex amplifiable probe hybridization
Abstract
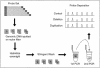
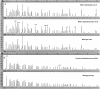
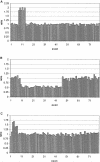
Duplications and deletions are known to cause a number of genetic disorders, yet technical difficulties and financial considerations mean that screening for these mutations, especially duplications, is often not performed. We have adapted multiplex amplifiable probe hybridization (MAPH) for the screening of the DMD gene, mutations in which cause Duchenne muscular dystrophy (DMD) and Becker muscular dystrophy. MAPH involves the quantitative recovery of specifically designed probes following hybridization to immobilized genomic DNA. We have engineered probes for each of the 79 exons of the DMD gene, and we analyzed them by using a 96-capillary sequencer. We screened 24 control individuals, 102 patients, and 23 potential carriers and detected a large number of novel rearrangements, especially small, one- and two-exon duplications. A duplication of exon 2 alone was the most frequently occurring mutation identified. Our analysis indicates that duplications occur in 6% of patients with DMD. The MAPH technique as modified here is simple, quick, and accurate; furthermore, it is based on existing technology (i.e., hybridization, PCR, and electrophoresis) and should not require new equipment. Together, these features should allow easy implementation in routine diagnostic laboratories. Furthermore, the methodology should be applicable to any genetic disease, it should be easily expandable to cover >200 probes, and its characteristics should facilitate high-throughput screening.
Figures
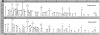

References
Electronic-Database Information
-
- BLAST, http://www.ncbi.nlm.nih.gov/BLAST/ (for nr and htgs databases)
-
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/
-
- Leiden Muscular Dystrophy Pages, http://www.dmd.nl/
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for DMD [MIM #310200])
References
-
- Bakker E, Van Broeckhoven C, Bonten EJ, Van De Vooren MJ, Veenema H, Van Hul W, van Ommen GJB, Vandenberghe A, Pearson PL (1987) Germline mosaicism and Duchenne muscular dystrophy mutations. Nature 329:554–556 - PubMed
-
- Beggs AH, Koenig M, Boyce FM, Kunkel LM (1990) Detection of 98% of DMD/BMD gene deletions by polymerase chain reaction. Hum Genet 86:45–48 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

