Genome properties of the diatom Phaeodactylum tricornutum
- PMID: 12114555
- PMCID: PMC166495
- DOI: 10.1104/pp.010713
Genome properties of the diatom Phaeodactylum tricornutum
Abstract
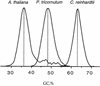
Diatoms are a ubiquitous class of microalgae of extreme importance for global primary productivity and for the biogeochemical cycling of minerals such as silica. However, very little is known about diatom cell biology or about their genome structure. For diatom researchers to take advantage of genomics and post-genomics technologies, it is necessary to establish a model diatom species. Phaeodactylum tricornutum is an obvious candidate because of its ease of culture and because it can be genetically transformed. Therefore, we have examined its genome composition by the generation of approximately 1,000 expressed sequence tags. Although more than 60% of the sequences could not be unequivocally identified by similarity to sequences in the databases, approximately 20% had high similarity with a range of genes defined functionally at the protein level. It is interesting that many of these sequences are more similar to animal rather than plant counterparts. Base composition at each codon position and GC content of the genome were compared with Arabidopsis, maize (Zea mays), and Chlamydomonas reinhardtii. It was found that distribution of GC within the coding sequences is as homogeneous in P. tricornutum as in Arabidopsis, but with a slightly higher GC content. Furthermore, we present evidence that the P. tricornutum genome is likely to be small (less than 20 Mb). Therefore, this combined information supports the development of this species as a model system for molecular-based studies of diatom biology. The nucleotide sequence data reported has been deposited in GenBank Nucleotide Sequence Database (dbEST section) under accession nos. BI306757 through BI307753.
Figures


References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Apt KE, Kroth-Pancic PG, Grossman AR. Stable nuclear transformation of the diatom Phaeodactylum tricornutum. Mol Gen Genet. 1997;252:572–579. - PubMed
-
- Asamizu E, Nakamura Y, Sato S, Tabata S. Generation of 7,37 non-redundant expressed sequence tags from a legume, Lotus japonicus. DNA Res. 2000;7:127–130. - PubMed
-
- Azam A, Paul J, Sehgal D, Prasad J, Bhattacharya S, Bhattacharya A. Identification of novel genes from Entamoeba histolytica by expressed sequence tag analysis. Gene. 1996;181:111–116. - PubMed
-
- Baldauf SL, Roger AJ, Wenk-Siefert I, Doolittle WF. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science. 2000;290:972–977. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

