PROLIFERATING INFLORESCENCE MERISTEM, a MADS-box gene that regulates floral meristem identity in pea
- PMID: 12114569
- PMCID: PMC166509
- DOI: 10.1104/pp.001677
PROLIFERATING INFLORESCENCE MERISTEM, a MADS-box gene that regulates floral meristem identity in pea
Abstract
SQUAMOSA and APETALA1 are floral meristem identity genes from snapdragon (Antirrhinum majus) and Arabidopsis, respectively. Here, we characterize the floral meristem identity mutation proliferating inflorescence meristem (pim) from pea (Pisum sativum) and show that it corresponds to a defect in the PEAM4 gene, a homolog of SQUAMOSA and APETALA1. The PEAM4 coding region was deleted in the pim-1 allele, and this deletion cosegregated with the pim-1 mutant phenotype. The pim-2 allele carried a nucleotide substitution at a predicted 5' splice site that resulted in mis-splicing of pim-2 mRNA. PCR products corresponding to unspliced and exon-skipped mRNA species were observed. The pim-1 and pim-2 mutations delayed floral meristem specification and altered floral morphology significantly but had no observable effect on vegetative development. These floral-specific mutant phenotypes and the restriction of PIM gene expression to flowers contrast with other known floral meristem genes in pea that additionally affect vegetative development. The identification of PIM provides an opportunity to compare pathways to flowering in species with different inflorescence architectures.
Figures

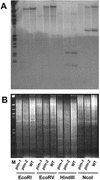
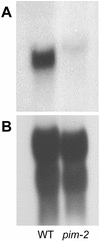



Similar articles
-
Analysis of PEAM4, the pea AP1 functional homologue, supports a model for AP1-like genes controlling both floral meristem and floral organ identity in different plant species.Plant J. 2001 Feb;25(4):441-51. doi: 10.1046/j.1365-313x.2001.00974.x. Plant J. 2001. PMID: 11260500
-
Virus-induced gene silencing of PEAM4 affects floral morphology by altering the expression pattern of PsSOC1a and PsPVP in pea.J Plant Physiol. 2014 Jan 15;171(2):148-53. doi: 10.1016/j.jplph.2013.08.010. Epub 2013 Nov 22. J Plant Physiol. 2014. PMID: 24331430
-
AGAMOUS-LIKE24 and SHORT VEGETATIVE PHASE determine floral meristem identity in Arabidopsis.Plant J. 2008 Dec;56(6):891-902. doi: 10.1111/j.1365-313X.2008.03648.x. Epub 2008 Aug 6. Plant J. 2008. PMID: 18694458
-
Reproductive meristem fates in Gerbera.J Exp Bot. 2006;57(13):3445-55. doi: 10.1093/jxb/erl181. Epub 2006 Oct 5. J Exp Bot. 2006. PMID: 17023564 Review.
-
Make hay when the sun shines: the role of MADS-box genes in temperature-dependant seasonal flowering responses.Plant Sci. 2011 Mar;180(3):447-53. doi: 10.1016/j.plantsci.2010.12.001. Epub 2010 Dec 14. Plant Sci. 2011. PMID: 21421391 Review.
Cited by
-
Modeling the Flowering Activation Motif during Vernalization in Legumes: A Case Study of M. trancatula.Life (Basel). 2023 Dec 23;14(1):26. doi: 10.3390/life14010026. Life (Basel). 2023. PMID: 38255642 Free PMC article.
-
Comparative transcriptional profiling provides insights into the evolution and development of the zygomorphic flower of Vicia sativa (Papilionoideae).PLoS One. 2013;8(2):e57338. doi: 10.1371/journal.pone.0057338. Epub 2013 Feb 21. PLoS One. 2013. PMID: 23437373 Free PMC article.
-
Plant Inflorescence Architecture: The Formation, Activity, and Fate of Axillary Meristems.Cold Spring Harb Perspect Biol. 2020 Jan 2;12(1):a034652. doi: 10.1101/cshperspect.a034652. Cold Spring Harb Perspect Biol. 2020. PMID: 31308142 Free PMC article. Review.
-
Dynamical Modeling of the Core Gene Network Controlling Transition to Flowering in Pisum sativum.Front Genet. 2021 Mar 11;12:614711. doi: 10.3389/fgene.2021.614711. eCollection 2021. Front Genet. 2021. PMID: 33777095 Free PMC article.
-
Altered regulation of flowering expands growth ranges and maximizes yields in major crops.Front Plant Sci. 2023 Jan 19;14:1094411. doi: 10.3389/fpls.2023.1094411. eCollection 2023. Front Plant Sci. 2023. PMID: 36743503 Free PMC article. Review.
References
-
- Berbel A, Navarro C, Ferrándiz C, Cañas LA, Madueño F, Beltrán J. Analysis of PEAM4, the pea AP1 functional homologue, supports a model for AP1-like genes controlling both floral meristem and floral organ identity in different plant species. Plant J. 2001;25:441–451. - PubMed
-
- Bowman JL, Alvarez J, Weigel D, Meyerowitz EM, Smyth DR. Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development. 1993;119:721–743.
-
- Bradley JM, Whitelam GC, Harberd NP. Impaired splicing of phytochrome B pre-mRNA in a novel phyB mutant of Arabidopsis. Plant Mol Biol. 1995;27:1133–1142. - PubMed
-
- Cremer F, Havelange A, Saedler H, Huijser P. Environmental control of flowering time in Antirrhinum majus. Physiol Plant. 1998;104:345–350.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

