Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene
- PMID: 12114572
- PMCID: PMC166512
- DOI: 10.1104/pp.011024
Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene
Abstract
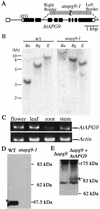
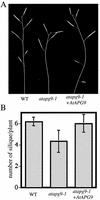
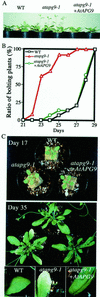
Autophagy is an intracellular process for vacuolar bulk degradation of cytoplasmic components. The molecular machinery responsible for yeast and mammalian autophagy has recently begun to be elucidated at the cellular level, but the role that autophagy plays at the organismal level has yet to be determined. In this study, a genome-wide search revealed significant conservation between yeast and plant autophagy genes. Twenty-five plant genes that are homologous to 12 yeast genes essential for autophagy were discovered. We identified an Arabidopsis mutant carrying a T-DNA insertion within AtAPG9, which is the only ortholog of yeast Apg9 in Arabidopsis (atapg9-1). AtAPG9 is transcribed in every wild-type organ tested but not in the atapg9-1 mutant. Under nitrogen or carbon-starvation conditions, chlorosis was observed earlier in atapg9-1 cotyledons and rosette leaves compared with wild-type plants. Furthermore, atapg9-1 exhibited a reduction in seed set when nitrogen starved. Even under nutrient growth conditions, bolting and natural leaf senescence were accelerated in atapg9-1 plants. Senescence-associated genes SEN1 and YSL4 were up-regulated in atapg9-1 before induction of senescence, unlike in wild type. All of these phenotypes were complemented by the expression of wild-type AtAPG9 in atapg9-1 plants. These results imply that autophagy is required for maintenance of the cellular viability under nutrient-limited conditions and for efficient nutrient use as a whole plant.
Figures





References
-
- Aubert S, Gout E, Bligny R, Marty-Mazars D, Barrieu F, Alabouvette J, Marty F, Douce R. Ultrastructural and biochemical characterization of autophagy in higher plant cells subjected to carbon deprivation: control by the supply of mitochondria with respiratory substrates. J Cell Biol. 1996;133:1251–1263. - PMC - PubMed
-
- Chen MH, Liu LF, Chen YR, Wu HK, Yu SM. Expression of alpha-amylases, carbohydrate metabolism, and autophagy in cultured rice cells is coordinately regulated by sugar nutrient. Plant J. 1994;6:625–636. - PubMed
-
- Chirgwin JJ, Przbyla AE, MacDonald RJ, Rutter WJ. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;18:5294–5299. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

