Phosphite, an analog of phosphate, suppresses the coordinated expression of genes under phosphate starvation
- PMID: 12114577
- PMCID: PMC166517
- DOI: 10.1104/pp.010835
Phosphite, an analog of phosphate, suppresses the coordinated expression of genes under phosphate starvation
Abstract
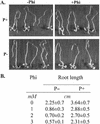
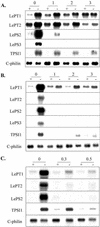
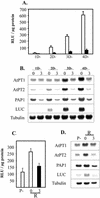
Phosphate (Pi) and its analog phosphite (Phi) are acquired by plants via Pi transporters. Although the uptake and mobility of Phi and Pi are similar, there is no evidence suggesting that plants can utilize Phi as a sole source of phosphorus. Phi is also known to interfere with many of the Pi starvation responses in plants and yeast (Saccharomyces cerevisiae). In this study, effects of Phi on plant growth and coordinated expression of genes induced by Pi starvation were analyzed. Phi suppressed many of the Pi starvation responses that are commonly observed in plants. Enhanced root growth and root to shoot ratio, a hallmark of Pi stress response, was strongly inhibited by Phi. The negative effects of Phi were not obvious in plants supplemented with Pi. The expression of Pi starvation-induced genes such as LePT1, LePT2, AtPT1, and AtPT2 (high-affinity Pi transporters); LePS2 (a novel acid phosphatase); LePS3 and TPSI1 (novel genes); and PAP1 (purple acid phosphatase) was suppressed by Phi in plants and cell cultures. Expression of luciferase reporter gene driven by the Pi starvation-induced AtPT2 promoter was also suppressed by Phi. These analyses showed that suppression of Pi starvation-induced genes is an early response to addition of Phi. These data also provide evidence that Phi interferes with gene expression at the level of transcription. Synchronized suppression of multiple Pi starvation-induced genes by Phi points to its action on the early molecular events, probably signal transduction, in Pi starvation response.
Figures


Similar articles
-
Suppression of the auxin response pathway enhances susceptibility to Phytophthora cinnamomi while phosphite-mediated resistance stimulates the auxin signalling pathway.BMC Plant Biol. 2014 Mar 20;14:68. doi: 10.1186/1471-2229-14-68. BMC Plant Biol. 2014. PMID: 24649892 Free PMC article.
-
Negative regulation of phosphate starvation-induced genes.Plant Physiol. 2001 Dec;127(4):1854-62. Plant Physiol. 2001. PMID: 11743129 Free PMC article.
-
Attenuation of phosphate starvation responses by phosphite in Arabidopsis.Plant Physiol. 2001 Nov;127(3):963-72. Plant Physiol. 2001. PMID: 11706178 Free PMC article.
-
Root architecture remodeling induced by phosphate starvation.Plant Signal Behav. 2011 Aug;6(8):1122-6. doi: 10.4161/psb.6.8.15752. Epub 2011 Aug 1. Plant Signal Behav. 2011. PMID: 21778826 Free PMC article. Review.
-
Phosphite: a novel P fertilizer for weed management and pathogen control.Plant Biotechnol J. 2017 Dec;15(12):1493-1508. doi: 10.1111/pbi.12803. Epub 2017 Sep 25. Plant Biotechnol J. 2017. PMID: 28776914 Free PMC article. Review.
Cited by
-
Acclimation responses of Arabidopsis thaliana to sustained phosphite treatments.J Exp Bot. 2013 Apr;64(6):1731-43. doi: 10.1093/jxb/ert037. Epub 2013 Feb 11. J Exp Bot. 2013. PMID: 23404904 Free PMC article.
-
Phosphorus nutrition of phosphorus-sensitive Australian native plants: threats to plant communities in a global biodiversity hotspot.Conserv Physiol. 2013 May 17;1(1):cot010. doi: 10.1093/conphys/cot010. eCollection 2013. Conserv Physiol. 2013. PMID: 27293594 Free PMC article.
-
Phosphate transport and homeostasis in Arabidopsis.Arabidopsis Book. 2002;1:e0024. doi: 10.1199/tab.0024. Epub 2002 Sep 30. Arabidopsis Book. 2002. PMID: 22303200 Free PMC article. No abstract available.
-
Differentiating phosphate-dependent and phosphate-independent systemic phosphate-starvation response networks in Arabidopsis thaliana through the application of phosphite.J Exp Bot. 2015 May;66(9):2501-14. doi: 10.1093/jxb/erv025. Epub 2015 Feb 19. J Exp Bot. 2015. PMID: 25697796 Free PMC article.
-
Phosphate availability regulates ethylene biosynthesis gene expression and protein accumulation in white clover (Trifolium repens L.) roots.Biosci Rep. 2016 Nov 17;36(6):e00411. doi: 10.1042/BSR20160148. Print 2016 Dec. Biosci Rep. 2016. PMID: 27737923 Free PMC article.
References
-
- Baldwin JC, Karthikeyan AS, Raghothama KG. Annual American Society of Plant Physiologists Meeting, July 24–28, Baltimore. Rockville, MD: American Society of Plant Physiologists; 1999. LePS3, a novel phosphate starvation induced gene in tomato (abstract no. 937) p. 190.
-
- Barchietto T, Saindrenan P, Bompeix G. Characterization of phosphonate uptake in two Phytophthora spp. and its inhibition by phosphate. Arch Microbiol. 1989;151:54–58.
-
- Barchietto T, Saindrenan P, Bompeix G. Physiological responses of Phytophthora citrophthora to a sub-inhibitory concentration of phosphonate. Pestic Biochem Physiol. 1992;42:151–166.
-
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

