Differential messenger RNA gradients in the unicellular alga Acetabularia acetabulum. Role of the cytoskeleton
- PMID: 12114594
- PMCID: PMC166534
- DOI: 10.1104/pp.010983
Differential messenger RNA gradients in the unicellular alga Acetabularia acetabulum. Role of the cytoskeleton
Abstract
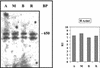
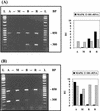
The unicellular green alga Acetabularia acetabulum has proven itself to be a superior model for studies of morphogenesis because of its large size and distinctive polar morphology. The giant cell forms an elongated tube (a stalk of up to 60 mm in length), which at its apical pole makes whorls of hairs, followed by one whorl of gametophores in the shape of a cap. At its basal pole, the cell extends into a rhizoid wherein the single nucleus is positioned. In this study, we have determined the level of specific messenger RNAs in the apical, middle, and basal regions using reverse transcriptase-PCR methodology. Four mRNA classes were distinguished: those that were uniformly distributed (small subunit of Rubisco, actin-1, ADP-glucose, centrin, and alpha- and beta-tubulin), those that expressed apical/basal (calmodulin-4) or basal/apical gradients (calmodulin-2 and a Ran-G protein), and those with development-specific patterns of distribution (mitogen-activated protein kinase, actin-2, and UDP-glucose-epimerase). Restoration of the apical/basal calmodulin-4 mRNA gradient after amputation of the apical region of the cell requires the nucleus and was abolished by cytochalasin D. Accumulation of actin-1 mRNA in the vicinity of the wound set by the amputation needs, likewise, the presence of the nucleus and was also inhibited by cytochalasin. This suggests that actin microfilaments of the cytoskeleton are involved in directed transport and/or anchoring of these mRNAs.
Figures







References
-
- Bachmann P, Zetsche K. A close temporal and spatial correlation between cell growth, cell wall synthesis and the activity of enzymes of mannan synthesis in Acetabularia mediterranea. Planta. 1979;145:331–337. - PubMed
-
- Bachs O, Agell N, Carafoli E. Calmodulin and calmodulin binding proteins in the nucleus. Cell Calcium. 1994;16:289–296. - PubMed
-
- Bashirullah A, Cooperstock RL, Lipshitz HD. RNA localization in development. Annu Rev Biochem. 1998;67:335–394. - PubMed
-
- Berger S, Kaever MJ. Dasycladales. Stuttgart, Germany: Georg Thieme Verlag; 1992.
-
- Berger S, Wittke W, Traub P. Occurrence of proteinaceous 10 nm filaments throughout the cytoplasm of algae of the order Dasycladales. Exp Cell Res. 1998;240:176–186. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

