Molecular cloning and characterization of genes for Shigella sonnei form I O polysaccharide: proposed biosynthetic pathway and stable expression in a live salmonella vaccine vector
- PMID: 12117952
- PMCID: PMC128211
- DOI: 10.1128/IAI.70.8.4414-4423.2002
Molecular cloning and characterization of genes for Shigella sonnei form I O polysaccharide: proposed biosynthetic pathway and stable expression in a live salmonella vaccine vector
Abstract
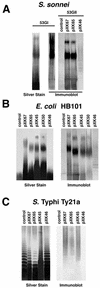
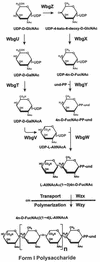
The gene region for biosynthesis of Shigella sonnei form I O polysaccharide (O-Ps) and flanking sequences, totaling >18 kb, was characterized by deletion analysis to define a minimal construct for development of Salmonella-based live vaccine vector strains. Lipopolysaccharide (LPS) expression and DNA sequence studies of plasmid deletion derivatives indicated form I O-Ps expression from a 12.3-kb region containing a putative promoter and 10 contiguous open reading frames (ORFs), one of which is the transposase of IS630. A detailed biosynthetic pathway, consistent with the predicted functions of eight of the nine essential ORFs and the form I O-Ps structure, is proposed. Further sequencing identified partial IS elements (i.e., IS91 and IS630) and wzz upstream of the form I coding region and a fragment of aqpZ and additional full or partial IS elements (i.e., IS629, IS91, and IS911) downstream of this region. The stability of plasmid-based form I O-Ps expression was greater from low-copy vectors than from high-copy vectors and was enhanced by deletion of the downstream IS91 from plasmid inserts. Both core-linked (i.e., LPS) and non-core-linked (i.e., capsule-like) surface expression of form I O-Ps were detected by Western blotting and silver staining of polyacrylamide gel electrophoresis-separated Shigella and Escherichia coli extracts. However, salmonellae, which have a core that is chemically dissimilar to that of shigellae, expressed only non-core-linked surface-associated form I O-Ps. Finally, attenuated Salmonella enterica serovar Typhi live vaccine vector candidates, containing minimal-sized form I operon constructs, elicited immune protection in mice against virulent S. sonnei challenge, thereby supporting the promise of live, oral vaccines for the prevention of shigellosis.
Figures

Similar articles
-
Stable expression of Shigella sonnei form I O-polysaccharide genes recombineered into the chromosome of live Salmonella oral vaccine vector Ty21a.Int J Med Microbiol. 2013 Apr;303(3):105-13. doi: 10.1016/j.ijmm.2013.01.001. Epub 2013 Mar 7. Int J Med Microbiol. 2013. PMID: 23474241
-
Stable Chromosomal Expression of Shigella flexneri 2a and 3a O-Antigens in the Live Salmonella Oral Vaccine Vector Ty21a.Clin Vaccine Immunol. 2017 Dec 5;24(12):e00181-17. doi: 10.1128/CVI.00181-17. Print 2017 Dec. Clin Vaccine Immunol. 2017. PMID: 29046309 Free PMC article.
-
Core-linked LPS expression of Shigella dysenteriae serotype 1 O-antigen in live Salmonella Typhi vaccine vector Ty21a: preclinical evidence of immunogenicity and protection.Vaccine. 2007 Aug 14;25(33):6167-75. doi: 10.1016/j.vaccine.2007.06.003. Epub 2007 Jun 26. Vaccine. 2007. PMID: 17629369
-
Shigellosis: the current status of vaccine development.Curr Opin Infect Dis. 2008 Jun;21(3):313-8. doi: 10.1097/QCO.0b013e3282f88b92. Curr Opin Infect Dis. 2008. PMID: 18448978 Review.
-
Inactivated and subunit vaccines to prevent shigellosis.Expert Rev Vaccines. 2009 Dec;8(12):1693-704. doi: 10.1586/erv.09.127. Expert Rev Vaccines. 2009. PMID: 19943764 Review.
Cited by
-
An O antigen capsule modulates bacterial pathogenesis in Shigella sonnei.PLoS Pathog. 2015 Mar 20;11(3):e1004749. doi: 10.1371/journal.ppat.1004749. eCollection 2015 Mar. PLoS Pathog. 2015. PMID: 25794007 Free PMC article.
-
Comparative Genomic and Phylogenetic Analysis of a Shiga Toxin Producing Shigella sonnei (STSS) Strain.Front Cell Infect Microbiol. 2017 May 30;7:229. doi: 10.3389/fcimb.2017.00229. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28611956 Free PMC article.
-
Two Novel Salmonella Bivalent Vaccines Confer Dual Protection against Two Salmonella Serovars in Mice.Front Cell Infect Microbiol. 2017 Sep 4;7:391. doi: 10.3389/fcimb.2017.00391. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28929089 Free PMC article.
-
Identification of the Vibrio vulnificus wbpP gene and evaluation of its role in virulence.Infect Immun. 2006 Jan;74(1):721-8. doi: 10.1128/IAI.74.1.721-728.2006. Infect Immun. 2006. PMID: 16369029 Free PMC article.
-
Anthrax protective antigen delivered by Salmonella enterica serovar Typhi Ty21a protects mice from a lethal anthrax spore challenge.Infect Immun. 2009 Apr;77(4):1475-82. doi: 10.1128/IAI.00828-08. Epub 2009 Jan 29. Infect Immun. 2009. PMID: 19179420 Free PMC article.
References
-
- Belanger, M., L. L. Burrows, and J. S. Lam. 1999. Functional analysis of genes responsible for the synthesis of the B-band O antigen of Pseudomonas aeruginosa serotype O6 lipopolysaccharide. Microbiology 145:3505-3521. - PubMed
-
- Black, R. E., M. M. Levine, M. L. Clements, G. Losonsky, D. Herrington, S. Berman, and S. B. Formal. 1987. Prevention of shigellosis by a Salmonella typhi-Shigella sonnei bivalent vaccine. J. Infect. Dis. 155:1260-1265. - PubMed
-
- Bolivar, F. 1978. Construction and characterization of new cloning vehicles. III. Derivatives of plasmid pBR322 carrying unique Eco RI sites for selection of Eco RI generated recombinant DNA molecules. Gene 4:121-136. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

