Genes for glycosylphosphatidylinositol toxin biosynthesis in Plasmodium falciparum
- PMID: 12117963
- PMCID: PMC128142
- DOI: 10.1128/IAI.70.8.4510-4522.2002
Genes for glycosylphosphatidylinositol toxin biosynthesis in Plasmodium falciparum
Abstract
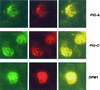
About 2.5 million people die of Plasmodium falciparum malaria every year. Fatalities are associated with systemic and organ-specific inflammation initiated by a parasite toxin. Recent studies show that glycosylphosphatidylinositol (GPI) functions as the dominant parasite toxin in the context of infection. GPIs also serve as membrane anchors for several of the most important surface antigens of parasite invasive stages. GPI anchoring is a complex posttranslational modification produced through the coordinated action of a multicomponent biosynthetic pathway. Here we present eight new genes of P. falciparum selected for encoding homologs of proteins essential for GPI synthesis: PIG-A, PIG-B, PIG-M, PIG-O, GPI1, GPI8, GAA-1, and DPM1. We describe the experimentally verified mRNA and predicted amino acid sequences and in situ localization of the gene products to the parasite endoplasmic reticulum. Moreover, we show preliminary evidence for the PIG-L and PIG-C genes. The biosynthetic pathway of the malaria parasite GPI offers potential targets for drug development and may be useful for studying parasite cell biology and the molecular basis for the pathophysiology of parasitic diseases.
Figures







Similar articles
-
The GPI1 homologue from Plasmodium falciparum complements a Saccharomyces cerevisiae GPI1 anchoring mutant.Mol Biochem Parasitol. 2002 Mar;120(1):73-81. doi: 10.1016/s0166-6851(01)00434-0. Mol Biochem Parasitol. 2002. PMID: 11849707
-
Complementation of essential yeast GPI mannosyltransferase mutations suggests a novel specificity for certain Trypanosoma and Plasmodium PigB proteins.PLoS One. 2014 Jan 29;9(1):e87673. doi: 10.1371/journal.pone.0087673. eCollection 2014. PLoS One. 2014. PMID: 24489949 Free PMC article.
-
PIG-C, one of the three human genes involved in the first step of glycosylphosphatidylinositol biosynthesis is a homologue of Saccharomyces cerevisiae GPI2.Biochem Biophys Res Commun. 1996 Sep 4;226(1):193-9. doi: 10.1006/bbrc.1996.1332. Biochem Biophys Res Commun. 1996. PMID: 8806613
-
Protein glycosylation in the malaria parasite.Parasitol Today. 1999 Apr;15(4):147-52. doi: 10.1016/s0169-4758(99)01412-x. Parasitol Today. 1999. PMID: 10322336 Review.
-
Enzymes and auxiliary factors for GPI lipid anchor biosynthesis and post-translational transfer to proteins.Bioessays. 2003 Apr;25(4):367-85. doi: 10.1002/bies.10254. Bioessays. 2003. PMID: 12655644 Review.
Cited by
-
Sugar activation and glycosylation in Plasmodium.Malar J. 2015 Oct 31;14:427. doi: 10.1186/s12936-015-0949-z. Malar J. 2015. PMID: 26520586 Free PMC article. Review.
-
Parasite Carbohydrate Vaccines.Front Cell Infect Microbiol. 2017 Jun 12;7:248. doi: 10.3389/fcimb.2017.00248. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28660174 Free PMC article. Review.
-
What Do We Know about Surface Proteins of Chicken Parasites Eimeria?Life (Basel). 2023 May 31;13(6):1295. doi: 10.3390/life13061295. Life (Basel). 2023. PMID: 37374079 Free PMC article. Review.
-
Malaria tolerance--for whom the cell tolls?Trends Parasitol. 2006 Aug;22(8):371-7. doi: 10.1016/j.pt.2006.06.002. Epub 2006 Jun 19. Trends Parasitol. 2006. PMID: 16784889 Free PMC article. Review.
-
Identification and functional analysis of Trypanosoma cruzi genes that encode proteins of the glycosylphosphatidylinositol biosynthetic pathway.PLoS Negl Trop Dis. 2013 Aug 8;7(8):e2369. doi: 10.1371/journal.pntd.0002369. eCollection 2013. PLoS Negl Trop Dis. 2013. PMID: 23951384 Free PMC article.
References
-
- Acosta-Serrano, A., S. Schenkman, N. Yoshida, A. Mehlert, J. M. Richardson, and M. A. J. Ferguson. 1995. The lipid structure of the glycosylphosphatidylinositol-anchored mucin-like sialic acid acceptors of Trypanosoma cruzi changes during parasite differentiation from epimastigotes to infective metacyclic trypomastigote forms. J. Biol. Chem. 270:27244-27253. - PubMed
-
- Baldauf, S. L., A. J. Roger, I. Wenk-Siefert, and W. F. Doolittle. 2000. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science 290:972-977. - PubMed
-
- Benachour, A., G. Sipos, I. Flury, F. Reggiori, E. Canivenc-Gansel, C. Vionnet, A. Conzelmann, and M. Benghezal. 1999. Deletion of GPI7, a yeast gene required for addition of a side chain to the glycosylphosphatidylinositol (GPI) core structure, affects GPI protein transport, remodeling, and cell wall integrity. J. Biol. Chem. 274:15251-15261. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

