Deductions about the number, organization, and evolution of genes in the tomato genome based on analysis of a large expressed sequence tag collection and selective genomic sequencing
- PMID: 12119366
- PMCID: PMC150698
- DOI: 10.1105/tpc.010478
Deductions about the number, organization, and evolution of genes in the tomato genome based on analysis of a large expressed sequence tag collection and selective genomic sequencing
Abstract
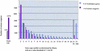
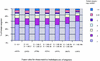
Analysis of a collection of 120,892 single-pass ESTs, derived from 26 different tomato cDNA libraries and reduced to a set of 27,274 unique consensus sequences (unigenes), revealed that 70% of the unigenes have identifiable homologs in the Arabidopsis genome. Genes corresponding to metabolism have remained most conserved between these two genomes, whereas genes encoding transcription factors are among the fastest evolving. The majority of the 10 largest conserved multigene families share similar copy numbers in tomato and Arabidopsis, suggesting that the multiplicity of these families may have occurred before the divergence of these two species. An exception to this multigene conservation was observed for the E8-like protein family, which is associated with fruit ripening and has higher copy number in tomato than in Arabidopsis. Finally, six BAC clones from different parts of the tomato genome were isolated, genetically mapped, sequenced, and annotated. The combined analysis of the EST database and these six sequenced BACs leads to the prediction that the tomato genome encodes approximately 35,000 genes, which are sequestered largely in euchromatic regions corresponding to less than one-quarter of the total DNA in the tomato nucleus.
Figures


References
-
- Adam, D. (2000). Now for the hard ones. Nature 408, 792–793. - PubMed
-
- Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. - PubMed
-
- Arumuganathan, K., and Earle, E.D. (1991). Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 9, 208–219.
-
- Bernatzky, R., and Tanksley, S.D. (1986). Majority of random cDNA clones correspond to single loci in the tomato genome. Mol. Gen. Genet. 203, 8–14.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

