Nucleocytoplasmic partitioning of the plant photoreceptors phytochrome A, B, C, D, and E is regulated differentially by light and exhibits a diurnal rhythm
- PMID: 12119373
- PMCID: PMC150705
- DOI: 10.1105/tpc.001156
Nucleocytoplasmic partitioning of the plant photoreceptors phytochrome A, B, C, D, and E is regulated differentially by light and exhibits a diurnal rhythm
Abstract
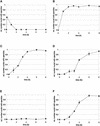
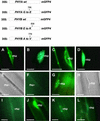
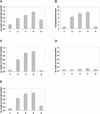
The phytochrome family of plant photoreceptors has a central role in the adaptation of plant development to changes in ambient light conditions. The individual phytochrome species regulate different or partly overlapping physiological responses. We generated transgenic Arabidopsis plants expressing phytochrome A to E:green fluorescent protein (GFP) fusion proteins to assess the biological role of intracellular compartmentation of these photoreceptors in light-regulated signaling. We show that all phytochrome:GFP fusion proteins were imported into the nuclei. Translocation of these photoreceptors into the nuclei was regulated differentially by light. Light-induced accumulation of phytochrome species in the nuclei resulted in the formation of speckles. The appearance of these nuclear structures exhibited distinctly different kinetics, wavelengths, and fluence dependence and was regulated by a diurnal rhythm. Furthermore, we demonstrate that the import of mutant phytochrome B:GFP and phytochrome A:GFP fusion proteins, shown to be defective in signaling in vivo, is regulated by light but is not accompanied by the formation of speckles. These results suggest that (1) the differential regulation of the translocation of phytochrome A to E into nuclei plays a role in the specification of functions, and (2) the appearance of speckles is a functional feature of phytochrome-regulated signaling.
Figures


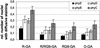

References
-
- Ang, L.-H., Chattopadhay, N.W., Oyama, T., Okada, K., Batschauer, A., and Deng, X.-W. (1998). Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol. Cell 1, 213–222. - PubMed
-
- Botto, J.F., Sanchez, R.A., and Casal, J.J. (1995). Role of phytochrome B in the induction of seed germination by light in Arabidopsis thaliana. J. Plant Physiol. 146, 307–312.
-
- Clack, T., Matthews, S., and Sharrock, R.A. (1994). The phytochrome apoprotein family in Arabidopsis is encoded by five genes: The sequence and expression of PHYD and PHYE. Plant Mol. Biol. 25, 413–417. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

