Ubiquitin ligase-associated protein SGT1 is required for host and nonhost disease resistance in plants
- PMID: 12119413
- PMCID: PMC125064
- DOI: 10.1073/pnas.152330599
Ubiquitin ligase-associated protein SGT1 is required for host and nonhost disease resistance in plants
Abstract
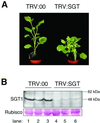
Homologues of the yeast ubiquitin ligase-associated protein SGT1 are required for disease resistance in plants mediated by nucleotide-binding site/leucine-rich repeat (NBS-LRR) proteins. Here, by silencing SGT1 in Nicotiana benthamiana, we extend these findings and demonstrate that SGT1 has an unexpectedly general role in disease resistance. It is required for resistance responses mediated by NBS-LRR and other R proteins in which pathogen-derived elicitors are recognized either inside or outside the host plant cell. A requirement also exists for SGT1 in nonhost resistance in which all known members of a host species are resistant against every characterized isolate of a pathogen. Our findings show that silencing SGT1 affects diverse types of disease resistance in plants and support the idea that R protein-mediated and nonhost resistance may involve similar mechanisms.
Figures


References
-
- Bonas U. & Lahaye, T. (2002) Curr. Opin. Microbiol. 5, 44-50. - PubMed
-
- Century K. S., Shapiro, A. D., Repetti, P. P., Dahlbeck, D., Holub, E. & Staskawicz, B. (1997) Science 278, 1963-1965. - PubMed
-
- Shirasu K., Lahaye, T., Tan, M.-W., Zhou, F., Azevedo, C. & Schulze-Lefert, P. (1999) Cell 99, 355-366. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

