Mapping the binding site of the neuroprotectant ifenprodil on NMDA receptors
- PMID: 12122058
- PMCID: PMC6757916
- DOI: 10.1523/JNEUROSCI.22-14-05955.2002
Mapping the binding site of the neuroprotectant ifenprodil on NMDA receptors
Abstract
Ifenprodil is a noncompetitive antagonist of NMDA receptors highly selective for the NMDA receptor 2B (NR2B) subunit. It is widely used as a pharmacological tool to discriminate subpopulations of NMDA receptors, and derivatives are currently being developed as candidate neuroprotectants. Despite numerous studies on the mechanism of action of ifenprodil on NMDA receptors, the structural determinants responsible for the subunit selectivity have not been identified. By combining functional studies on recombinant NMDA receptors and biochemical studies on isolated domains, we now show that ifenprodil binds to the N-terminal leucine/isoleucine/valine-binding protein (LIVBP)-like domain of NR2B. In this domain, several residues, both hydrophilic and hydrophobic, were found to control ifenprodil inhibition. Their location in a modeled three-dimensional structure suggests that ifenprodil binds in the cleft of the LIVBP-like domain of NR2B by a mechanism (Venus-flytrap) resembling that of the binding of Zn on the LIVBP-like domain of NR2A. These results reinforce the proposal that the LIVBP-like domains of NMDA receptors, and possibly of other ionotropic glutamate receptors, bind modulatory ligands. Moreover, they identify the LIVBP-like domain of the NR2B subunit as a promising therapeutic target and provide a framework for designing structurally novel NR2B-selective antagonists.
Figures





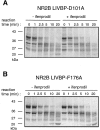
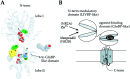
References
-
- Armstrong N, Sun Y, Chen GQ, Gouaux E. Structure of a glutamate-receptor ligand-binding core in complex with kainate. Nature. 1998;395:913–917. - PubMed
-
- Armstrong N, Gouaux E. Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: crystal structures of the GluR2 ligand binding core. Neuron. 2000;28:165–181. - PubMed
-
- Ayalon G, Stern-Bach Y. Functional assembly of AMPA and kainate receptors is mediated by several discrete protein-protein interactions. Neuron. 2001;31:103–113. - PubMed
-
- Carter C, Benavides J, Legendre P, Vincent JD, Noel F, Thuret F, Lloyd KG, Arbilla S, Zivkovic B, MacKenzie ET, Scatton B, Langer SZ. Ifenprodil and SL 82.0715 as cerebral anti-ischemic agents: II. Evidence for N-methyl-d-aspartate receptor antagonist properties. J Pharmacol Exp Ther. 1988;247:1222–1232. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
