Defining signal thresholds in DNA microarrays: exemplary application for invasive cancer
- PMID: 12123529
- PMCID: PMC117791
- DOI: 10.1186/1471-2164-3-19
Defining signal thresholds in DNA microarrays: exemplary application for invasive cancer
Abstract
Background: Genome-wide or application-targeted microarrays containing a subset of genes of interest have become widely used as a research tool with the prospect of diagnostic application. Intrinsic variability of microarray measurements poses a major problem in defining signal thresholds for absent/present or differentially expressed genes. Most strategies have used fold-change threshold values, but variability at low signal intensities may invalidate this approach and it does not provide information about false-positives and false negatives.
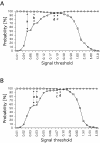
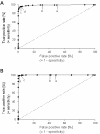
Results: We introduce a method to filter false-positives and false-negatives from DNA microarray experiments. This is achieved by evaluating a set of positive and negative controls by receiver operating characteristic (ROC) analysis. As an advantage of this approach, users may define thresholds on the basis of sensitivity and specificity considerations. The area under the ROC curve allows quality control of microarray hybridizations. This method has been applied to custom made microarrays developed for the analysis of invasive melanoma derived tumor cells. It demonstrated that ROC analysis yields a threshold with reduced missclassified genes in microarray experiments.
Conclusions: Provided that a set of appropriate positive and negative controls is included on the microarray, ROC analysis obviates the inherent problem of arbitrarily selecting threshold levels in microarray experiments. The proposed method is applicable to both custom made and commercially available DNA microarrays and will help to improve the reliability of predictions from DNA microarray experiments.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

