Imprinted genes: identification by chromosome rearrangements and post-genomic strategies
- PMID: 12127776
- PMCID: PMC2814292
- DOI: 10.1016/s0168-9525(02)02708-7
Imprinted genes: identification by chromosome rearrangements and post-genomic strategies
Abstract
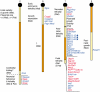
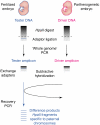
Imprinted genes are differentially expressed from the maternally and paternally inherited alleles. Accordingly, inheritance of both copies of an imprinted chromosome or region from a single parent leads to the mis-expression of the imprinted genes present in the selected region. Strains of mice with reciprocal and Robertsonian chromosomal translocations or mice with engineered chromosomal rearrangements can be used to produce progeny where both copies of a chromosomal region are inherited from one parent. In combination with systematic differential expression and methylation-based approaches, these mice can be used to identify novel imprinted genes. Advances in genome sequencing and computer-based technologies have facilitated this approach to finding imprinted genes.
Figures
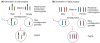

Similar articles
-
Parent-of-origin effects on gene expression and DNA methylation in the maize endosperm.Plant Cell. 2011 Dec;23(12):4221-33. doi: 10.1105/tpc.111.092668. Epub 2011 Dec 23. Plant Cell. 2011. PMID: 22198147 Free PMC article.
-
Genomic imprinting of the IGF2R/AIR locus is conserved between bovines and mice.Theriogenology. 2022 Mar 1;180:121-129. doi: 10.1016/j.theriogenology.2021.12.013. Epub 2021 Dec 20. Theriogenology. 2022. PMID: 34971973
-
Methylation screening of reciprocal genome-wide UPDs identifies novel human-specific imprinted genes.Hum Mol Genet. 2011 Aug 15;20(16):3188-97. doi: 10.1093/hmg/ddr224. Epub 2011 May 18. Hum Mol Genet. 2011. PMID: 21593219
-
Maternal regulation of chromosomal imprinting in animals.Chromosoma. 2019 Jun;128(2):69-80. doi: 10.1007/s00412-018-00690-5. Epub 2019 Feb 5. Chromosoma. 2019. PMID: 30719566 Free PMC article. Review.
-
Imprinting of the mouse Igf2r gene depends on an intronic CpG island.Mol Cell Endocrinol. 1998 May 25;140(1-2):9-14. doi: 10.1016/s0303-7207(98)00022-7. Mol Cell Endocrinol. 1998. PMID: 9722161 Review.
Cited by
-
Investigating parent of origin effects in studies of type 2 diabetes and obesity.Curr Diabetes Rev. 2008 Nov;4(4):329-39. doi: 10.2174/157339908786241179. Curr Diabetes Rev. 2008. PMID: 18991601 Free PMC article. Review.
-
Genomic imprinting in mammals: emerging themes and established theories.PLoS Genet. 2006 Nov 24;2(11):e147. doi: 10.1371/journal.pgen.0020147. PLoS Genet. 2006. PMID: 17121465 Free PMC article. Review.
-
Next-Generation Sequencing Techniques Reveal that Genomic Imprinting Is Absent in Day-Old Gallus gallus domesticus Brains.PLoS One. 2015 Jul 10;10(7):e0132345. doi: 10.1371/journal.pone.0132345. eCollection 2015. PLoS One. 2015. PMID: 26161857 Free PMC article.
-
The Prader-Willi syndrome murine imprinting center is not involved in the spatio-temporal transcriptional regulation of the Necdin gene.BMC Genet. 2005 Jan 5;6:1. doi: 10.1186/1471-2156-6-1. BMC Genet. 2005. PMID: 15634360 Free PMC article.
-
Phenotype analysis by MUC2, MUC5AC, MUC6, and CD10 expression in Epstein-Barr virus-associated gastric carcinoma.J Gastroenterol. 2006 Aug;41(8):733-9. doi: 10.1007/s00535-006-1841-y. J Gastroenterol. 2006. PMID: 16988760
References
-
- Cattanach BM, Beechey CV. Genomic imprinting in the mouse: possible final analysis. In: Reik W, Surani A, editors. Genomic Imprinting: Frontiers in Molecular Biology. Vol. 18. IRLPress; 1997. pp. 118–145.
-
- Mills AA, Bradley A. From mouse to man: generating megabase chromosome rearrangements. Trends Genet. 2001;17:331–339. - PubMed
-
- Cleary MA, et al. Disruption of an imprinted gene cluster by a targeted chromosomal tranlocation in mice. Nat. Genet. 2001;29:78–82. - PubMed
-
- Cattanach BM, Kirk M. Differential activity of maternally and paternally derived chromosome regions in mice. Nature. 1985;315:496–498. - PubMed
-
- Cattanach BM, Beechey CV. Genomic imprinting in the mouse: possible final analysis. In: Reik W, Surani A, editors. Genomic Imprinting: Frontiers in Molecular Biology. Vol. 18. IRL Press; 1997. pp. 118–145.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

