Imprinted genes: identification by chromosome rearrangements and post-genomic strategies
- PMID: 12127776
- PMCID: PMC2814292
- DOI: 10.1016/s0168-9525(02)02708-7
Imprinted genes: identification by chromosome rearrangements and post-genomic strategies
Abstract
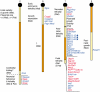
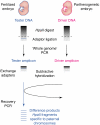
Imprinted genes are differentially expressed from the maternally and paternally inherited alleles. Accordingly, inheritance of both copies of an imprinted chromosome or region from a single parent leads to the mis-expression of the imprinted genes present in the selected region. Strains of mice with reciprocal and Robertsonian chromosomal translocations or mice with engineered chromosomal rearrangements can be used to produce progeny where both copies of a chromosomal region are inherited from one parent. In combination with systematic differential expression and methylation-based approaches, these mice can be used to identify novel imprinted genes. Advances in genome sequencing and computer-based technologies have facilitated this approach to finding imprinted genes.
Figures
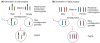

References
-
- Cattanach BM, Beechey CV. Genomic imprinting in the mouse: possible final analysis. In: Reik W, Surani A, editors. Genomic Imprinting: Frontiers in Molecular Biology. Vol. 18. IRLPress; 1997. pp. 118–145.
-
- Mills AA, Bradley A. From mouse to man: generating megabase chromosome rearrangements. Trends Genet. 2001;17:331–339. - PubMed
-
- Cleary MA, et al. Disruption of an imprinted gene cluster by a targeted chromosomal tranlocation in mice. Nat. Genet. 2001;29:78–82. - PubMed
-
- Cattanach BM, Kirk M. Differential activity of maternally and paternally derived chromosome regions in mice. Nature. 1985;315:496–498. - PubMed
-
- Cattanach BM, Beechey CV. Genomic imprinting in the mouse: possible final analysis. In: Reik W, Surani A, editors. Genomic Imprinting: Frontiers in Molecular Biology. Vol. 18. IRL Press; 1997. pp. 118–145.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

