A nonsense mutation in the fibrillin-1 gene of a Marfan syndrome patient induces NMD and disrupts an exonic splicing enhancer
- PMID: 12130535
- PMCID: PMC186389
- DOI: 10.1101/gad.997502
A nonsense mutation in the fibrillin-1 gene of a Marfan syndrome patient induces NMD and disrupts an exonic splicing enhancer
Abstract
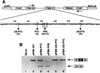
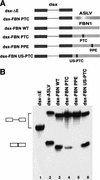
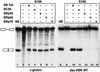
A nonsense mutation in the fibrillin-1 (FBN1) gene of a Marfan syndrome (MFS) patient induces in-frame exon skipping of FBN1 exon 51. We present evidence, based on both in vivo and in vitro experiments, that the skipping of this exon is due to the disruption of an SC35-dependent splicing enhancer within exon 51. In addition, this nonsense mutation induces nonsense-mediated decay (NMD), which degrades the normally spliced mRNA in the patient's cells. In contrast to NMD, skipping of FBN1 exon 51 does not require translation.
Figures

Comment in
-
NASty effects on fibrillin pre-mRNA splicing: another case of ESE does it, but proposals for translation-dependent splice site choice live on.Genes Dev. 2002 Jul 15;16(14):1743-53. doi: 10.1101/gad.1014502. Genes Dev. 2002. PMID: 12130534 Review. No abstract available.
References
-
- Carter MS, Doskow J, Morris P, Li S, Nhim RP, Sandstedt S, Wilkinson MF. A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J Biol Chem. 1995;270:28995–29003. - PubMed
-
- Dietz HC, Kendzior RJ., Jr Maintenance of an open reading frame as an additional level of scrutiny during splice site selection. Nat Genet. 1994;8:183–188. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
