Multiple transmembrane amino acid requirements suggest a highly specific interaction between the bovine papillomavirus E5 oncoprotein and the platelet-derived growth factor beta receptor
- PMID: 12134002
- PMCID: PMC155141
- DOI: 10.1128/jvi.76.16.7976-7986.2002
Multiple transmembrane amino acid requirements suggest a highly specific interaction between the bovine papillomavirus E5 oncoprotein and the platelet-derived growth factor beta receptor
Abstract
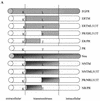
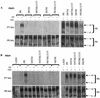
The bovine papillomavirus E5 protein activates the cellular platelet-derived growth factor beta receptor (PDGFbetaR) tyrosine kinase in a ligand-independent manner. Evidence suggests that the small transmembrane E5 protein homodimerizes and physically interacts with the transmembrane domain of the PDGFbetaR, thereby inducing constitutive dimerization and activation of this receptor. Amino acids in the receptor previously found to be required for the PDGFbetaR-E5 interaction are a transmembrane Thr513 and a juxtamembrane Lys499. Here, we sought to determine if these are the only two receptor amino acids required for an interaction with the E5 protein. Substitution of large portions of the PDGFbetaR transmembrane domain indicated that additional amino acids in both the amino and carboxyl halves of the receptor transmembrane domain are required for a productive interaction with the E5 protein. Indeed, individual amino acid substitutions in the receptor transmembrane domain identified roles for the extracellular proximal transmembrane residues in the interaction. These data suggest that multiple amino acids within the transmembrane domain of the PDGFbetaR are required for a stable interaction with the E5 protein. These may be involved in direct protein-protein contacts or may support the proper transmembrane alpha-helical conformation for optimal positioning of the primary amino acid requirements.
Figures





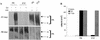
Similar articles
-
The bovine papillomavirus E5 protein and the PDGF beta receptor: it takes two to tango.Virology. 2009 Feb 20;384(2):345-51. doi: 10.1016/j.virol.2008.09.033. Epub 2008 Nov 6. Virology. 2009. PMID: 18990418 Free PMC article. Review.
-
Molecular examination of the transmembrane requirements of the platelet-derived growth factor beta receptor for a productive interaction with the bovine papillomavirus E5 oncoprotein.J Biol Chem. 2002 Dec 6;277(49):47149-59. doi: 10.1074/jbc.M209582200. Epub 2002 Sep 25. J Biol Chem. 2002. PMID: 12351659
-
Identification of amino acids in the transmembrane and juxtamembrane domains of the platelet-derived growth factor receptor required for productive interaction with the bovine papillomavirus E5 protein.J Virol. 1997 Oct;71(10):7318-27. doi: 10.1128/JVI.71.10.7318-7327.1997. J Virol. 1997. PMID: 9311809 Free PMC article.
-
The platelet-derived growth factor beta receptor as a target of the bovine papillomavirus E5 protein.Cytokine Growth Factor Rev. 2000 Dec;11(4):283-93. doi: 10.1016/s1359-6101(00)00012-5. Cytokine Growth Factor Rev. 2000. PMID: 10959076 Review.
-
A single amino acid substitution converts a transmembrane protein activator of the platelet-derived growth factor β receptor into an inhibitor.J Biol Chem. 2013 Sep 20;288(38):27273-27286. doi: 10.1074/jbc.M113.470054. Epub 2013 Aug 1. J Biol Chem. 2013. PMID: 23908351 Free PMC article.
Cited by
-
The bovine papillomavirus E5 protein and the PDGF beta receptor: it takes two to tango.Virology. 2009 Feb 20;384(2):345-51. doi: 10.1016/j.virol.2008.09.033. Epub 2008 Nov 6. Virology. 2009. PMID: 18990418 Free PMC article. Review.
-
Compensatory mutants of the bovine papillomavirus E5 protein and the platelet-derived growth factor β receptor reveal a complex direct transmembrane interaction.J Virol. 2013 Oct;87(20):10936-45. doi: 10.1128/JVI.01475-13. Epub 2013 Aug 7. J Virol. 2013. PMID: 23926343 Free PMC article.
-
The E5 proteins.Virology. 2013 Oct;445(1-2):99-114. doi: 10.1016/j.virol.2013.05.006. Epub 2013 May 31. Virology. 2013. PMID: 23731971 Free PMC article. Review.
-
Viral miniproteins.Annu Rev Microbiol. 2014;68:21-43. doi: 10.1146/annurev-micro-091313-103727. Epub 2014 Apr 10. Annu Rev Microbiol. 2014. PMID: 24742054 Free PMC article. Review.
-
Two transmembrane dimers of the bovine papillomavirus E5 oncoprotein clamp the PDGF β receptor in an active dimeric conformation.Proc Natl Acad Sci U S A. 2017 Aug 29;114(35):E7262-E7271. doi: 10.1073/pnas.1705622114. Epub 2017 Aug 14. Proc Natl Acad Sci U S A. 2017. PMID: 28808001 Free PMC article.
References
-
- Bergman, P., M. Ustav, J. Sedman, J. Moreno-Lopez, B. Vennstrom, and U. Pettersson. 1988. The E5 gene of bovine papillomavirus type 1 is sufficient for complete oncogenic transformation of mouse fibroblasts. Oncogene 2:453-459. - PubMed
-
- Burkhardt, A., M. Willingham, C. Gay, K. T. Jeang, and R. Schlegel. 1989. The E5 oncoprotein of bovine papillomavirus is oriented asymmetrically in Golgi and plasma membranes. Virology 170:334-339. - PubMed
-
- Claesson-Welsh, L. 1994. Platelet-derived growth factor receptor signals. J. Biol. Chem. 269:32023-32026. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

