Given the opportunity, the Sendai virus RNA-dependent RNA polymerase could as well enter its template internally
- PMID: 12134003
- PMCID: PMC155117
- DOI: 10.1128/jvi.76.16.7987-7995.2002
Given the opportunity, the Sendai virus RNA-dependent RNA polymerase could as well enter its template internally
Abstract
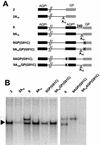
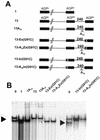
The negative-stranded RNA viral genome is an RNA-protein complex of helicoidal symmetry, resistant to nonionic detergent and high salt, in which the RNA is protected from RNase digestion. The 15,384 nucleotides of the Sendai virus genome are bound to 2,564 subunits of the N protein, each interacting with six nucleotides so tightly that the bases are poorly accessible to soluble reagents. With such a uniform structure, the question of template recognition by the viral RNA polymerase has been raised. In a previous study, the N-phase context has been proposed to be crucial for this recognition, a notion referring to the importance of the position in which the nucleotides interact with the N protein. The N-phase context ruled out the role of the template 3'-OH congruence, a feature resulting from the obedience to the rule of six that implies the precise interaction of the last six 3'-OH nucleotides with the last N protein. The N-phase context then allows prediction of the recognition by the RNA polymerase of a replication promoter sequence even if internally positioned, a promoter which normally lies at the template extremity. In this study, with template minireplicons bearing tandem replication promoters separated by intervening sequences, we present data that indeed show that initiation of RNA synthesis takes place at the internal promoter. This internal initiation can best be interpreted as the result of the polymerase entering the template at the internal promoter. In this way, the data are consistent with the importance of the N-phase context in template recognition. Moreover, by introducing between the two promoters a stretch of 10 A residues which represent a barrier for RNA synthesis, we found that the ability of the RNA polymerase to cross this barrier depends on the type of replication promoter, strong or weak, that the RNA polymerase starts on, a sign that the RNA polymerase may be somehow imprinted in its activity by the nature of the promoter on which it starts synthesis.
Figures






Similar articles
-
Moieties in an RNA promoter specifically recognized by a viral RNA-dependent RNA polymerase.Proc Natl Acad Sci U S A. 1998 Sep 29;95(20):11613-8. doi: 10.1073/pnas.95.20.11613. Proc Natl Acad Sci U S A. 1998. PMID: 9751714 Free PMC article.
-
"Rule of six": how does the Sendai virus RNA polymerase keep count?J Virol. 2001 May;75(10):4506-18. doi: 10.1128/JVI.75.10.4506-4518.2001. J Virol. 2001. PMID: 11312321 Free PMC article.
-
Chemical modification of nucleotide bases and mRNA editing depend on hexamer or nucleoprotein phase in Sendai virus nucleocapsids.RNA. 2002 Aug;8(8):1056-67. doi: 10.1017/s1355838202029977. RNA. 2002. PMID: 12212849 Free PMC article.
-
Viral DNA polymerase scanning and the gymnastics of Sendai virus RNA synthesis.Virology. 2004 Jan 20;318(2):463-73. doi: 10.1016/j.virol.2003.10.031. Virology. 2004. PMID: 15015496 Review.
-
Structural insights into RNA synthesis by the influenza virus transcription-replication machine.Virus Res. 2017 Apr 15;234:103-117. doi: 10.1016/j.virusres.2017.01.013. Epub 2017 Jan 20. Virus Res. 2017. PMID: 28115197 Review.
Cited by
-
Evidence that the respiratory syncytial virus polymerase is recruited to nucleotides 1 to 11 at the 3' end of the nucleocapsid and can scan to access internal signals.J Virol. 2005 Sep;79(17):11311-22. doi: 10.1128/JVI.79.17.11311-11322.2005. J Virol. 2005. PMID: 16103183 Free PMC article.
-
Evidence that the polymerase of respiratory syncytial virus initiates RNA replication in a nontemplated fashion.Proc Natl Acad Sci U S A. 2010 Jun 1;107(22):10226-31. doi: 10.1073/pnas.0913065107. Epub 2010 May 17. Proc Natl Acad Sci U S A. 2010. PMID: 20479224 Free PMC article.
-
RNA polymerase II-controlled expression of antigenomic RNA enhances the rescue efficacies of two different members of the Mononegavirales independently of the site of viral genome replication.J Virol. 2006 Jun;80(12):5708-15. doi: 10.1128/JVI.02389-05. J Virol. 2006. PMID: 16731909 Free PMC article.
-
Bunyamwera virus can repair both insertions and deletions during RNA replication.RNA. 2010 Jun;16(6):1138-45. doi: 10.1261/rna.1962010. Epub 2010 Apr 29. RNA. 2010. PMID: 20430858 Free PMC article.
-
Active borna disease virus polymerase complex requires a distinct nucleoprotein-to-phosphoprotein ratio but no viral X protein.J Virol. 2003 Nov;77(21):11781-9. doi: 10.1128/jvi.77.21.11781-11789.2003. J Virol. 2003. PMID: 14557662 Free PMC article.
References
-
- Boyer, J.-C., and A.-L. Haenni. 1994. Infectious transcripts and cDNA clones of RNA viruses. Virology 198:415-426. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

