Bacteriophage PM2 has a protein capsid surrounding a spherical proteinaceous lipid core
- PMID: 12134022
- PMCID: PMC155172
- DOI: 10.1128/jvi.76.16.8169-8178.2002
Bacteriophage PM2 has a protein capsid surrounding a spherical proteinaceous lipid core
Abstract
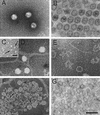
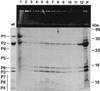
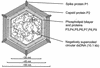
The marine double-stranded DNA (dsDNA) bacteriophage PM2, studied since 1968, is the type organism of the family Corticoviridae, infecting two gram-negative Pseudoalteromonas species. The virion contains a membrane underneath an icosahedral protein capsid composed of two structural proteins. The purified major capsid protein, P2, appears as a trimer, and the receptor binding protein, P1, appears as a monomer. The C-terminal part of P1 is distal and is responsible for receptor binding activity. The rest of the structural proteins are associated with the internal phospholipid membrane enclosing the viral genome. This internal particle is designated the lipid core. The overall structural organization of phage PM2 resembles that of dsDNA bacteriophage PRD1, the type organism of the family TECTIVIRIDAE:
Figures
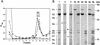



Similar articles
-
Penetration of membrane-containing double-stranded-DNA bacteriophage PM2 into Pseudoalteromonas hosts.J Bacteriol. 2004 Aug;186(16):5342-54. doi: 10.1128/JB.186.16.5342-5354.2004. J Bacteriol. 2004. PMID: 15292135 Free PMC article.
-
ICTV Virus Taxonomy Profile: Corticoviridae.J Gen Virol. 2017 May;98(5):888-889. doi: 10.1099/jgv.0.000795. J Gen Virol. 2017. PMID: 28581380 Free PMC article.
-
Preliminary crystallographic analysis of the major capsid protein P2 of the lipid-containing bacteriophage PM2.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2005 Aug 1;61(Pt 8):762-5. doi: 10.1107/S174430910502141X. Epub 2005 Jul 30. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2005. PMID: 16511151 Free PMC article.
-
Lipid-containing viruses: bacteriophage PRD1 assembly.Adv Exp Med Biol. 2012;726:365-77. doi: 10.1007/978-1-4614-0980-9_16. Adv Exp Med Biol. 2012. PMID: 22297522 Review.
-
From lows to highs: using low-resolution models to phase X-ray data.Acta Crystallogr D Biol Crystallogr. 2013 Nov;69(Pt 11):2257-65. doi: 10.1107/S0907444913022336. Epub 2013 Oct 18. Acta Crystallogr D Biol Crystallogr. 2013. PMID: 24189238 Free PMC article. Review.
Cited by
-
The structure of the NTPase that powers DNA packaging into Sulfolobus turreted icosahedral virus 2.J Virol. 2013 Aug;87(15):8388-98. doi: 10.1128/JVI.00831-13. Epub 2013 May 22. J Virol. 2013. PMID: 23698307 Free PMC article.
-
Penetration of membrane-containing double-stranded-DNA bacteriophage PM2 into Pseudoalteromonas hosts.J Bacteriol. 2004 Aug;186(16):5342-54. doi: 10.1128/JB.186.16.5342-5354.2004. J Bacteriol. 2004. PMID: 15292135 Free PMC article.
-
Ultrastructure and Viral Metagenome of Bacteriophages from an Anaerobic Methane Oxidizing Methylomirabilis Bioreactor Enrichment Culture.Front Microbiol. 2016 Nov 8;7:1740. doi: 10.3389/fmicb.2016.01740. eCollection 2016. Front Microbiol. 2016. PMID: 27877158 Free PMC article.
-
Jorvik: A membrane-containing phage that will likely found a new family within Vinavirales.iScience. 2023 Sep 29;26(11):108104. doi: 10.1016/j.isci.2023.108104. eCollection 2023 Nov 17. iScience. 2023. PMID: 37867962 Free PMC article.
-
Bacteriophage Capsid Modification by Genetic and Chemical Methods.Bioconjug Chem. 2021 Mar 17;32(3):466-481. doi: 10.1021/acs.bioconjchem.1c00018. Epub 2021 Mar 4. Bioconjug Chem. 2021. PMID: 33661607 Free PMC article. Review.
References
-
- Akutsu, H., H. Satake, and R. M. Franklin. 1980. Phosphorus nuclear magnetic resonance studies on the lipid-containing bacteriophage PM2. Biochemistry 19:5264-5270. - PubMed
-
- Armour, G. A., and G. J. Brewer. 1990. Membrane morphogenesis from cloned fragments of bacteriophage PM2 DNA that contain the sp6.6 gene. FASEB J. 4:1488-1493. - PubMed
-
- Bamford, D. H., J. Caldentey, and J. K. H. Bamford. 1995. Bacteriophage PRD1: a broad host range dsDNA tectivirus with an internal membrane. Adv. Virus Res. 45:281-319. - PubMed
-
- Bamford, D. H., and L. Mindich. 1980. Electron microscopy of cells infected with nonsense mutants of bacteriophage φ6. Virology 107:222-228. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

