Molecular architecture of adenovirus DNA polymerase and location of the protein primer
- PMID: 12134025
- PMCID: PMC155156
- DOI: 10.1128/jvi.76.16.8200-8207.2002
Molecular architecture of adenovirus DNA polymerase and location of the protein primer
Abstract
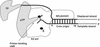
Adenovirus (Ad) DNA polymerase (pol) belongs to the distinct subclass of the polalpha family of DNA pols that employs the precursor terminal protein (pTP) as primer. Ad pol forms a stable heterodimer with this primer, and together, they bind specifically to the core origin in order to start replication. After initiation of Ad replication, the resulting pTP-trinucleotide intermediate jumps back and pTP starts to dissociate. Compared to free Ad pol, the pTP-pol complex shows reduced polymerase and exonuclease activities, but the reason for this is not understood. Furthermore, the interaction domains between these proteins have not been defined and the contribution of each protein to origin binding is unclear. To address these questions, we used oligonucleotides with a translocation block and show here that pTP binds at the entrance of the primer binding groove of Ad pol, thereby explaining the decreased synthetic activities of the pTP-pol complex and providing insight into how pTP primes Ad replication. Employing an exonuclease-deficient mutant polymerase, we further show that the polymerase and exonuclease active sites of Ad pol are spatially distinct and that the exonuclease activity of Ad pol is located at the N-terminal part of the protein. In addition, by probing the distances between both active sites and the surface of Ad pol, we show that Ad pol binds a DNA region of 14 to 15 nucleotides. Based on these results, a model for binding of the pTP-pol complex at the origin of replication is proposed.
Figures
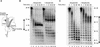
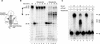


Similar articles
-
Properties of the adenovirus DNA polymerase.J Biol Chem. 1984 Aug 10;259(15):9487-95. J Biol Chem. 1984. PMID: 6540263
-
Dissociation of the protein primer and DNA polymerase after initiation of adenovirus DNA replication.J Biol Chem. 1997 Sep 26;272(39):24617-23. doi: 10.1074/jbc.272.39.24617. J Biol Chem. 1997. PMID: 9305930
-
DNA binding properties of the adenovirus DNA replication priming protein pTP.Nucleic Acids Res. 2003 Jun 15;31(12):3274-86. doi: 10.1093/nar/gkg405. Nucleic Acids Res. 2003. PMID: 12799455 Free PMC article.
-
Mechanism of DNA replication in eukaryotic cells: cellular host factors stimulating adenovirus DNA replication.Gene. 1999 Aug 5;236(1):1-12. doi: 10.1016/s0378-1119(99)00249-8. Gene. 1999. PMID: 10433960 Review.
-
Adenovirus DNA replication: protein priming, jumping back and the role of the DNA binding protein DBP.Curr Top Microbiol Immunol. 2003;272:187-211. doi: 10.1007/978-3-662-05597-7_7. Curr Top Microbiol Immunol. 2003. PMID: 12747551 Review.
Cited by
-
Adenovirus type 5 DNA binding protein stimulates binding of DNA polymerase to the replication origin.J Virol. 2003 Jan;77(2):915-22. doi: 10.1128/jvi.77.2.915-922.2003. J Virol. 2003. PMID: 12502807 Free PMC article.
-
Interregional Coevolution Analysis Revealing Functional and Structural Interrelatedness between Different Genomic Regions in Human Mastadenovirus D.J Virol. 2015 Jun;89(12):6209-17. doi: 10.1128/JVI.00515-15. Epub 2015 Apr 1. J Virol. 2015. PMID: 25833048 Free PMC article.
-
High Fidelity Deep Sequencing Reveals No Effect of ATM, ATR, and DNA-PK Cellular DNA Damage Response Pathways on Adenovirus Mutation Rate.Viruses. 2019 Oct 11;11(10):938. doi: 10.3390/v11100938. Viruses. 2019. PMID: 31614688 Free PMC article.
-
Adenovirus DNA replication.Cold Spring Harb Perspect Biol. 2013 Mar 1;5(3):a013003. doi: 10.1101/cshperspect.a013003. Cold Spring Harb Perspect Biol. 2013. PMID: 23388625 Free PMC article. Review.
-
Selective modification of adenovirus replication can be achieved through rational mutagenesis of the adenovirus type 5 DNA polymerase.J Virol. 2012 Oct;86(19):10484-93. doi: 10.1128/JVI.00739-12. Epub 2012 Jul 18. J Virol. 2012. PMID: 22811532 Free PMC article.
References
-
- Bernad, A., L. Blanco, J. M. Lazaro, G. Martin, and M. Salas. 1989. A conserved 3′-5′ exonuclease active site in prokaryotic and eukaryotic DNA polymerases. Cell 59:219-228. - PubMed
-
- Blanco, L., and M. Salas. 1995. Mutational analysis of bacteriophage phi 29 DNA polymerase. Methods Enzymol. 262:283-294. - PubMed
-
- Blasco, M. A., J. Mendez, J. M. Lazaro, L. Blanco, and M. Salas. 1995. Primer terminus stabilization at the phi 29 DNA polymerase active site. Mutational analysis of conserved motif KXY. J. Biol. Chem. 270:2735-2740. - PubMed
-
- Brenkman, A. B., M. R. Heideman, V. Truniger, M. Salas, and P. C. Der Vliet. 2001. The (I/Y)XGG motif of adenovirus DNA polymerase affects template DNA binding and the transition from initiation to elongation. J. Biol. Chem. 276:29846-29853. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

