The long noncoding region of the human parainfluenza virus type 1 f gene contributes to the read-through transcription at the m-f gene junction
- PMID: 12134030
- PMCID: PMC155142
- DOI: 10.1128/jvi.76.16.8244-8251.2002
The long noncoding region of the human parainfluenza virus type 1 f gene contributes to the read-through transcription at the m-f gene junction
Abstract
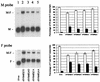
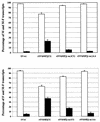
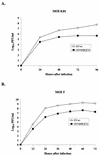
Sendai virus (SV) and human parainfluenza virus type 1 (hPIV1) have genomes consisting of nonsegmented negative-sense RNA in which the six genes are separated by well-conserved intergenic (IG) sequences and transcriptional start (S) and end signals. In hPIV1-infected cells, transcriptional termination at the M-F gene junction is ineffective; a large number of M-F read-through transcripts are produced (T. Bousse, T. Takimoto, K. G. Murti, and A. Portner, Virology 232:44-52, 1997). In contrast, few M-F read-through transcripts are detected in SV-infected cells. Sequence analysis indicated that the hPIV1 IG and S sequences in the M-F junction differ from those of SV. Furthermore, the hPIV1 F gene contains an unusually long noncoding sequence. To identify the cis-acting elements that prevent transcriptional termination at the M-F junction, we rescued recombinant SV (rSVhMFjCG) in which its M-F gene junction was replaced by that of hPIV1. Cells infected with rSVhMFjCG produced an abundance of M-F read-through transcripts; this result indicated that the hPIV1 M-F junction is responsible for inefficient termination. When one or both of the IG and S sites in rSVhMFjCG were replaced by those of SV, the efficiency of transcriptional termination increased but not to the level observed in wild-type SV-infected cells. Deletion of most of the long noncoding region of the hPIV1 F gene in rSVhMFjCG in addition to the mutations in IG and S signals resulted in efficient termination that was equivalent to the level observed in wild-type virus-infected cells. Therefore, the long noncoding sequence of the hPIV1 F gene contains cis-acting element(s) that affects transcriptional termination. Our evaluation of the effect of inefficient transcriptional termination on viral replication in culture revealed that cells infected with rSVhMFjCG produced less F protein than cells infected with wild-type SV and that assembly of the recombinant SV in culture was less efficient. These phenotypes seem to be responsible for the extended survival of mice infected with rSVhMFjCG.
Figures


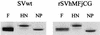
References
-
- Bousse, T., T. Takimoto, W. L. Gorman, T. Takahashi, and A. Portner. 1994. Regions on the hemagglutinin-neuraminidase protein of human parainfluenza virus type 1 and Sendai virus important for membrane fusion. Virology 204:506-514. - PubMed
-
- Bousse, T., T. Takimoto, T. Matrosovich, and A. Portner. 2001. Two regions of the P protein are required to be active with L protein for human parainfluenza virus type 1 RNA polymerase activity. Virology 283:306-314. - PubMed
-
- Bousse, T., T. Takimoto, K. G. Murti, and A. Portner. 1997. Elevated expression of the human parainfluenza virus type 1 F gene downregulates HN expression. Virology 232:44-52. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

