Ordered nucleation and spreading of silenced chromatin in Saccharomyces cerevisiae
- PMID: 12134062
- PMCID: PMC117306
- DOI: 10.1091/mbc.e02-03-0175
Ordered nucleation and spreading of silenced chromatin in Saccharomyces cerevisiae
Abstract
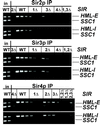
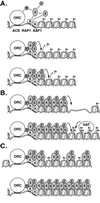
In Saccharomyces cerevisiae, silencing at the HM loci depends on Sir proteins, which are structural components of silenced chromatin. To explore the structure and assembly of silenced chromatin, the associations of Sir proteins with sequences across the HMR locus were examined by chromatin immunoprecipitation. In wild-type cells, Sir2p, Sir3p, and Sir4p were spread throughout and coincident with the silenced region at HMR. Sir1p, in contrast, associated only with the HMR-E silencer, consistent with its role in establishment but not maintenance of silencing. Sir4p was required for the association of other Sir proteins with silencers. In contrast, in the absence of Sir2p or Sir3p, partial assemblies of Sir proteins could form at silencers, where Sir protein assembly began. Spreading across HMR required Sir2p and Sir3p, as well as the deacetylase activity of Sir2p. These data support a model for the spreading of silenced chromatin involving cycles of nucleosome deacetylation by Sir2p followed by recruitment of additional Sir2p, Sir3p, and Sir4p to the newly deacetylated nucleosome. This model suggests mechanisms for boundary formation, and for maintenance and inheritance of silenced chromatin. The principles are generalizable to other types of heritable chromatin states.
Figures






References
-
- Abraham J, Nasmyth KA, Strathern JN, Klar AJ, Hicks JB. Regulation of mating-type information in yeast. Negative control requiring sequences both 5′ and 3′ to the regulated region. J Mol Biol. 1984;176:307–331. - PubMed
-
- Axelrod AR. Role of a Cell-Cycle Gene in Transcriptional Silencing. Ph.D. Thesis. Berkeley, CA: University of California, Berkeley; 1991.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

