PC phosphorylation increases the ability of AFAP-110 to cross-link actin filaments
- PMID: 12134071
- PMCID: PMC117315
- DOI: 10.1091/mbc.e01-12-0148
PC phosphorylation increases the ability of AFAP-110 to cross-link actin filaments
Abstract
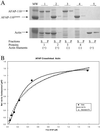
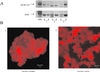
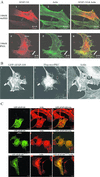
The actin filament-associated protein and Src-binding partner, AFAP-110, is an adaptor protein that links signaling molecules to actin filaments. AFAP-110 binds actin filaments directly and multimerizes through a leucine zipper motif. Cellular signals downstream of Src(527F) can regulate multimerization. Here, we determined recombinant AFAP-110 (rAFAP-110)-bound actin filaments cooperatively, through a lateral association. We demonstrate rAFAP-110 has the capability to cross-link actin filaments, and this ability is dependent on the integrity of the carboxy terminal actin binding domain. Deletion of the leucine zipper motif or PKC phosphorylation affected AFAP-110's conformation, which correlated with changes in multimerization and increased the capability of rAFAP-110 to cross-link actin filaments. AFAP-110 is both a substrate and binding partner of PKC. On PKC activation, stress filament organization is lost, motility structures form, and AFAP-110 colocalizes strongly with motility structures. Expression of a deletion mutant of AFAP-110 that is unable to bind PKC blocked the effect of PMA on actin filaments. We hypothesize that upon PKC activation, AFAP-110 can be cooperatively recruited to newly forming actin filaments, like those that exist in cell motility structures, and that PKC phosphorylation effects a conformational change that may enable AFAP-110 to promote actin filament cross-linking at the cell membrane.
Figures





References
-
- Baisden JM, Qian Y, Zot HG, Flynn DC. The actin filament associated protein, AFAP-110, is an adaptor protein that modulates changes in actin filament integrity. Oncogene. 2001a;20:6435–6447. - PubMed
-
- Baisden JM, Gatesman AS, Cherezova L, Jiang BH, Flynn DC. The intrinsic ability of AFAP-110 to alter actin filament integrity is linked with its ability to also activate cellular tyrosine kinases. Oncogene. 2001b;20:6607–6616. - PubMed
-
- Blikstad I, Markey F, Carlsson L, Persson T, Lindberg U. Selective assay of monomeric and filamentous actin in cell extracts using inhibition of deoxyribonuclease I. Cell. 1978;55:935–943. - PubMed
-
- Bourguignon LY, Zhu H, Shao L, Chen YW. CD44 interaction with c-Src kinase promotes cortactin-mediated cytoskeleton function and hyaluronic acid (HA)-dependent ovarian tumor cell migration. J Biol Chem. 2001;276:7327–7336. - PubMed
-
- Boyle WJ, van der Geer P, Hunter T. Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol. 1991;201:110–149. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

