A conserved Drosophila transportin-serine/arginine-rich (SR) protein permits nuclear import of Drosophila SR protein splicing factors and their antagonist repressor splicing factor 1
- PMID: 12134081
- PMCID: PMC117325
- DOI: 10.1091/mbc.e02-02-0102
A conserved Drosophila transportin-serine/arginine-rich (SR) protein permits nuclear import of Drosophila SR protein splicing factors and their antagonist repressor splicing factor 1
Abstract
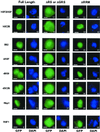
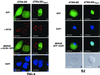
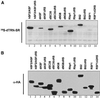
Members of the highly conserved serine/arginine-rich (SR) protein family are nuclear factors involved in splicing of metazoan mRNA precursors. In mammals, two nuclear import receptors, transportin (TRN)-SR1 and TRN-SR2, are responsible for targeting SR proteins to the nucleus. Distinctive features in the nuclear localization signal between Drosophila and mammalian SR proteins prompted us to examine the mechanism by which Drosophila SR proteins and their antagonist repressor splicing factor 1 (RSF1) are imported into nucleus. Herein, we report the identification and characterization of a Drosophila importin beta-family protein (dTRN-SR), homologous to TRN-SR2, that specifically interacts with both SR proteins and RSF1. dTRN-SR has a broad localization in the cytoplasm and the nucleus, whereas an N-terminal deletion mutant colocalizes with SR proteins in nuclear speckles. Far Western experiments established that the RS domain of SR proteins and the GRS domain of RSF1 are required for the direct interaction with dTRN-SR, an interaction that can be modulated by phosphorylation. Using the yeast model system in which nuclear import of Drosophila SR proteins and RSF1 is impaired, we demonstrate that complementation with dTRN-SR is sufficient to target these proteins to the nucleus. Together, the results imply that the mechanism by which SR proteins are imported to the nucleus is conserved between Drosophila and humans.
Figures

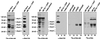
References
-
- Aitchison JD, Blobel G, Rout MP. Kap104p: a karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science. 1996;274:624–627. - PubMed
-
- Amrein H, Hedley ML, Maniatis T. The role of specific protein-RNA and protein-protein interactions in positive and negative control of pre-mRNA splicing by Transformer 2. Cell. 1994;76:735–746. - PubMed
-
- Blencowe BJ. Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem. 2000;25:106–110. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

