Bone morphogenetic protein-4-induced activation of Xretpos is mediated by Smads and Olf-1/EBF associated zinc finger (OAZ)
- PMID: 12136093
- PMCID: PMC135757
- DOI: 10.1093/nar/gkf437
Bone morphogenetic protein-4-induced activation of Xretpos is mediated by Smads and Olf-1/EBF associated zinc finger (OAZ)
Abstract
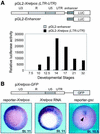
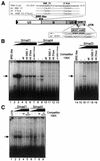
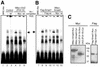
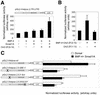
We have previously isolated Xretpos, a novel family of long terminal repeat (LTR)-retrotransposons in Xenopus laevis, whose transcript is restricted to ventro-posterior-specific regions and induced by bone morphogenetic protein-4 (BMP-4) signaling. To explore the molecular mechanism of the transcriptional regulation, we identified and characterized Xretpos promoter regions consisting of LTRs and a 5'-untranslated region. We demonstrated that this promoter region contains all the necessary regulatory elements for the spatial and temporal expression of XRETPOS: Sequence analysis of the Xretpos promoter revealed multiple Smad-binding elements and Olf-1/EBF-associated zinc finger (OAZ) binding sites similar to BMP-4 response element, which were identified and proved to be required for BMP-4 induction in the Xvent2 promoter. We further demonstrated that Smads and OAZ proteins bind to their response elements in the promoter and these bindings are essential for the BMP-4-induced activation of the Xretpos promoter. Furthermore, we showed that the endogenous expression of Xretpos protein indeed occurred and was temporally regulated and BMP-4-inducible during the early Xenopus development. Finally, overexpression and partial loss-of-function study revealed that Xretpos has a posterio-ventralizing activity. Together, our results place Xretpos downstream of BMP-4 and provide evidence for the conserved mechanism of transcriptional regulation of the BMP-4 target genes.
Figures





References
-
- Massagué J., Blain,S.W. and Lo,R.S. (2000) TGFβ signaling in growth control, cancer, and heritable disorders. Cell, 103, 295–309. - PubMed
-
- Hill C.S., (2001) TGF-β signalling pathways in early Xenopus development. Curr. Opin. Genet. Dev., 11, 533–540. - PubMed
-
- Whitman M., (1998) Smads and early developmental signaling by the TGFβ superfamily. Genes Dev., 12, 2445–2462. - PubMed

