The role of template-primer in protection of reverse transcriptase from thermal inactivation
- PMID: 12136094
- PMCID: PMC135738
- DOI: 10.1093/nar/gkf417
The role of template-primer in protection of reverse transcriptase from thermal inactivation
Abstract
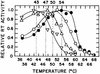
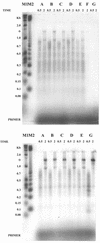
We compared the thermal stabilities of wild-type recombinant avian myeloblastosis virus (AMV) and Moloney murine leukemia virus (M-MLV) reverse transcriptase (RT) with those of mutants of the recombinant enzymes lacking RNase H activity. They differed in resistance to thermal inactivation at elevated temperatures in the presence of an RNA/DNA template-primer. RNase H-minus RTs retained the ability to efficiently synthesize cDNA at much higher temperatures. We show that the structure of the template-primer has a critical bearing on protection of RT from thermal inactivation. RT RNase H activity rapidly alters the structure of the template-primer to forms less tightly bound by RT and thus less able to protect the enzyme at elevated temperatures. We also found that when comparing wild-type or mutant AMV RT with the respective M-MLV RT, the avian enzymes retained more DNA synthetic activity at elevated temperatures than murine RTs. Enzyme, template-primer interaction again played the most significant role in producing these differences. AMV RT binds much tighter to template- primer and has a much greater tendency to remain bound during cDNA synthesis than M-MLV RT and therefore is better protected from heat inactivation.
Figures


References
-
- Prasad V.R., (1993) Genetic analysis of retroviral reverse transcriptase structure and function. In Skalka,A.M. and Goff,S.P. (eds), Reverse Transcriptases. CSHL Press, Plainview, NY, pp. 135–162.
-
- Moelling K., (1974) Characterization of reverse transcriptase and RNase H from Friend-murine leukemia virus. Virology, 62, 46–59. - PubMed
-
- Kotewicz M.L., D’Alessio,J.M., Driftmier,K.M., Blodgett,K.P. and Gerard,G.F. (1985) Cloning and overexpression of Moloney murine leukemia virus reverse transcriptase in Escherichia coli. Gene, 35, 249–258. - PubMed
-
- Roth M.J., Tanese,N. and Goff,S.P. (1985) Purification and characterization of murine retroviral reverse transcriptase expressed in Escherichia coli. J. Biol. Chem., 260, 9326–9335. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

