Real-time monitoring of rolling-circle amplification using a modified molecular beacon design
- PMID: 12136114
- PMCID: PMC135767
- DOI: 10.1093/nar/gnf065
Real-time monitoring of rolling-circle amplification using a modified molecular beacon design
Abstract
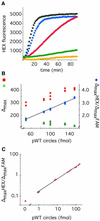
We describe a method to monitor rolling-circle replication of circular oligonucleotides in dual-color and in real-time using molecular beacons. The method can be used to study the kinetics of the polymerization reaction and to amplify and quantify circularized oligonucleotide probes in a rolling-circle amplification (RCA) reaction. Modified molecular beacons were made of 2'-O-Me-RNA to prevent 3' exonucleolytic degradation by the polymerase used. Moreover, the complement of one of the stem sequences of the molecular beacon was included in the RCA products to avoid fluorescence quenching due to inter-molecular hybridization of neighboring molecular beacons hybridizing to the concatemeric polymerization product. The method allows highly accurate quantification of circularized DNA over a broad concentration range by relating the signal from the test DNA circle to an internal reference DNA circle reporting in a distinct fluorescence color.
Figures
Similar articles
-
Coupled rolling circle amplification loop-mediated amplification for rapid detection of short DNA sequences.Biotechniques. 2008 Sep;45(3):275-80. doi: 10.2144/000112910. Biotechniques. 2008. PMID: 18778251
-
Real-time monitoring of branched rolling-circle DNA amplification with peptide nucleic acid beacon.Anal Biochem. 2004 Dec 15;335(2):326-9. doi: 10.1016/j.ab.2004.07.022. Anal Biochem. 2004. PMID: 15556572 No abstract available.
-
Signal amplification of padlock probes by rolling circle replication.Nucleic Acids Res. 1998 Nov 15;26(22):5073-8. doi: 10.1093/nar/26.22.5073. Nucleic Acids Res. 1998. PMID: 9801302 Free PMC article.
-
Toward improved biochips based on rolling circle amplification--influences of the microenvironment on the fluorescence properties of labeled DNA oligonucleotides.Ann N Y Acad Sci. 2008;1130:287-92. doi: 10.1196/annals.1430.022. Ann N Y Acad Sci. 2008. PMID: 18596361 Review.
-
Amplification of circularizable probes for the detection of target nucleic acids and proteins.Clin Chim Acta. 2006 Jan;363(1-2):61-70. doi: 10.1016/j.cccn.2005.05.039. Epub 2005 Aug 24. Clin Chim Acta. 2006. PMID: 16122721 Review.
Cited by
-
Development of a panel of recombinase polymerase amplification assays for detection of biothreat agents.J Clin Microbiol. 2013 Apr;51(4):1110-7. doi: 10.1128/JCM.02704-12. Epub 2013 Jan 23. J Clin Microbiol. 2013. PMID: 23345286 Free PMC article.
-
Simultaneous Detection of Different Zika Virus Lineages via Molecular Computation in a Point-of-Care Assay.Viruses. 2018 Dec 14;10(12):714. doi: 10.3390/v10120714. Viruses. 2018. PMID: 30558136 Free PMC article.
-
Integration of microbead DNA handling with optomagnetic detection in rolling circle amplification assays.Mikrochim Acta. 2019 Jul 11;186(8):528. doi: 10.1007/s00604-019-3636-x. Mikrochim Acta. 2019. PMID: 31297615
-
Enzymatic signal amplification of molecular beacons for sensitive DNA detection.Nucleic Acids Res. 2008 Apr;36(6):e36. doi: 10.1093/nar/gkn033. Epub 2008 Feb 27. Nucleic Acids Res. 2008. PMID: 18304948 Free PMC article.
-
Single-copy detection of somatic variants from solid and liquid biopsy.Sci Rep. 2021 Mar 16;11(1):6068. doi: 10.1038/s41598-021-85545-3. Sci Rep. 2021. PMID: 33727644 Free PMC article.
References
-
- Heid C.A., Stevens,J., Livak,K.J. and Williams,P.M. (1996) Real time quantitative PCR. Genome Res., 6, 986–994. - PubMed
-
- Tyagi S., and Kramer,F.R. (1996) Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol., 14, 303–308. - PubMed
-
- Li J.J., Fang,X., Schuster,S.M. and Tan,W. (2000) Molecular beacons: a novel approach to detect protein–DNA interactions. Angew. Chem. Int. Ed., 39, 1049–1052. - PubMed
-
- Lizardi P.M., Huang,X., Zhu,Z., Bray-Ward,P., Thomas,D.C. and Ward,D.C. (1998) Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nature Genet., 19, 225–232. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

