Multiplex SNP genotyping in pooled DNA samples by a four-colour microarray system
- PMID: 12136118
- PMCID: PMC135771
- DOI: 10.1093/nar/gnf069
Multiplex SNP genotyping in pooled DNA samples by a four-colour microarray system
Abstract
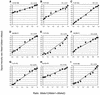
We selected 125 candidate single nucleotide polymorphisms (SNPs) in genes belonging to the human type 1 interferon (IFN) gene family and the genes coding for proteins in the main type 1 IFN signalling pathway by screening databases and by in silico comparison of DNA sequences. Using quantitative analysis of pooled DNA samples by solid-phase mini-sequencing, we found that only 20% of the candidate SNPs were polymorphic in the Finnish and Swedish populations. To allow more effective validation of candidate SNPs, we developed a four-colour microarray-based mini-sequencing assay for multiplex, quantitative allele frequency determination in pooled DNA samples. We used cyclic mini-sequencing reactions with primers carrying 5'-tag sequences, followed by capture of the products on microarrays by hybridisation to complementary tag oligonucleotides. Standard curves prepared from mixtures of known amounts of SNP alleles demonstrate the applicability of the system to quantitative analysis, and showed that for about half of the tested SNPs the limit of detection for the minority allele was below 5%. The microarray-based genotyping system established here is universally applicable for genotyping and quantification of any SNP, and the validated system for SNPs in type 1 IFN-related genes should find many applications in genetic studies of this important immunoregulatory pathway.
Figures



References
-
- Sachidanandam R., Weissman,D., Schmidt,S.C., Kakol,J.M., Stein,L.D., Marth,G., Sherry,S., Mullikin,J.C., Mortimore,B.J., Willey,D.L. et al. (2001) A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature, 409, 928–933. - PubMed
-
- Douabin-Gicquel V., Soriano,N., Ferran,H., Wojcik,F., Palierne,E., Tamim,S., Jovelin,T., McKie,A.T., Le Gall,J.Y., David,V. et al. (2001) Identification of 96 single nucleotide polymorphisms in eight genes involved in iron metabolism: efficiency of bioinformatic extraction compared with a systematic sequencing approach. Hum. Genet., 109, 393–401. - PubMed
-
- Marth G., Yeh,R., Minton,M., Donaldson,R., Li,Q., Duan,S., Davenport,R., Miller,R.D. and Kwok,P.Y. (2001) Single-nucleotide polymorphisms in the public domain: how useful are they? Nature Genet., 27, 371–372. - PubMed
-
- Kruglyak L., (1999) Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nature Genet., 22, 139–144. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

