Transcriptional regulation of the mdm2 oncogene by p53 requires TRRAP acetyltransferase complexes
- PMID: 12138177
- PMCID: PMC133988
- DOI: 10.1128/MCB.22.16.5650-5661.2002
Transcriptional regulation of the mdm2 oncogene by p53 requires TRRAP acetyltransferase complexes
Abstract
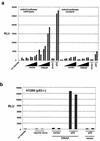
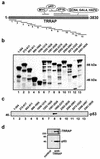
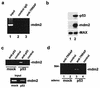
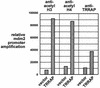
The p53 tumor suppressor regulates the cellular response to genetic damage through its function as a sequence-specific transcription factor. Among the most well-characterized transcriptional targets of p53 is the mdm2 oncogene. Activation of mdm2 is critical in the p53 pathway because the mdm2 protein marks p53 for proteosome-mediated degradation, thereby providing a negative-feedback loop. Here we show that the ATM-related TRRAP protein functionally cooperates with p53 to activate mdm2 transcription. TRRAP is a component of several multiprotein acetyltransferase complexes implicated in both transcriptional regulation and DNA repair. In support of a role for these complexes in mdm2 expression, we show that transactivation of the mdm2 gene is augmented by pharmacological inhibition of cellular deacetylases. In vitro analysis demonstrates that p53 directly binds to a TRRAP domain previously shown to be an activator docking site. Furthermore, transfection of cells with antisense TRRAP blocks p53-dependent transcription of mdm2. Finally, using chromatin immunoprecipitation, we demonstrate direct p53-dependent recruitment of TRRAP to the mdm2 promoter, followed by increased histone acetylation. These findings suggest a model in which p53 directly recruits a TRRAP/acetyltransferase complex to the mdm2 gene to activate transcription. In addition, this study defines a novel biochemical mechanism utilized by the p53 tumor suppressor to regulate gene expression.
Figures





References
-
- Arata, Y., M. Fujita, K. Ohtani, S. Kijima, and J. Y. Kato. 2000. Cdk2-dependent and -independent pathways in E2F-mediated S phase induction. J. Biol. Chem. 275:6337-6345. - PubMed
-
- Barak, Y., E. Gottlieb, T. Juven-Gershon, and M. Oren. 1994. Regulation of mdm2 expression by p53: alternative promoters produce transcripts with nonidentical translation potential. Genes Dev. 8:1739-1749. - PubMed
-
- Barlev, N. A., L. Liu, N. H. Chehab, K. Mansfield, K. G. Harris, T. D. Halazonetis, and S. L. Berger. 2001. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol. Cell 8:1243-1254. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
