Involvement of replicative polymerases, Tel1p, Mec1p, Cdc13p, and the Ku complex in telomere-telomere recombination
- PMID: 12138180
- PMCID: PMC133992
- DOI: 10.1128/MCB.22.16.5679-5687.2002
Involvement of replicative polymerases, Tel1p, Mec1p, Cdc13p, and the Ku complex in telomere-telomere recombination
Abstract
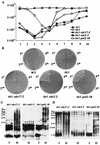
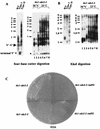
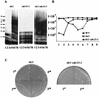
Telomere maintenance is required for chromosome stability, and telomeres are typically replicated by the action of the reverse transcriptase telomerase. In both tumor and yeast cells that lack telomerase, telomeres are maintained by an alternative recombination mechanism. Genetic studies have led to the identification of DNA polymerases, cell cycle checkpoint proteins, and telomere binding proteins involved in the telomerase pathway. However, how these proteins affect telomere-telomere recombination has not been identified to date. Using an assay to trace the in vivo recombinational products throughout the course of survivor development, we show here that three major replicative polymerases, alpha, delta, and epsilon, play roles in telomere-telomere recombination and that each causes different effects and phenotypes when they as well as the telomerase are defective. Polymerase delta appears to be the main activity for telomere extension, since neither type I nor type II survivors arising via telomere-telomere recombination were seen in its absence. The frequency of type I versus type II is altered in the polymerase alpha and epsilon mutants relative to the wild type. Each prefers to develop a particular type of survivor. Moreover, type II recombination is mediated by the cell cycle checkpoint proteins Tel1 and Mec1, and telomere-telomere recombination is regulated by telomere binding protein Cdc13 and the Ku complex. Together, our results suggest that coordination between DNA replication machinery, DNA damage signaling, DNA recombination machinery, and the telomere protein-DNA complex allows telomere recombination to repair telomeric ends in the absence of telomerase.
Figures

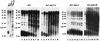

Similar articles
-
Telomere maintenance is dependent on activities required for end repair of double-strand breaks.Curr Biol. 1998 May 21;8(11):657-60. doi: 10.1016/s0960-9822(98)70253-2. Curr Biol. 1998. PMID: 9635193
-
Altering telomere structure allows telomerase to act in yeast lacking ATM kinases.Curr Biol. 2001 Aug 21;11(16):1240-50. doi: 10.1016/s0960-9822(01)00391-8. Curr Biol. 2001. PMID: 11525738
-
Cdc13 cooperates with the yeast Ku proteins and Stn1 to regulate telomerase recruitment.Mol Cell Biol. 2000 Nov;20(22):8397-408. doi: 10.1128/MCB.20.22.8397-8408.2000. Mol Cell Biol. 2000. PMID: 11046137 Free PMC article.
-
Telomeres--unsticky ends.Science. 1998 Sep 18;281(5384):1818-9. doi: 10.1126/science.281.5384.1818. Science. 1998. PMID: 9776685 Review. No abstract available.
-
Sir-Ku-itous routes to make ends meet.Cell. 1999 Jun 25;97(7):829-32. doi: 10.1016/s0092-8674(00)80795-3. Cell. 1999. PMID: 10399911 Review. No abstract available.
Cited by
-
Genistein suppresses the proliferation of telomerase-negative cells.Food Sci Nutr. 2016 May 26;5(2):197-204. doi: 10.1002/fsn3.382. eCollection 2017 Mar. Food Sci Nutr. 2016. PMID: 28265354 Free PMC article.
-
An mre11 mutation that promotes telomere recombination and an efficient bypass of senescence.Genetics. 2010 Jul;185(3):761-70. doi: 10.1534/genetics.110.117598. Epub 2010 Apr 26. Genetics. 2010. PMID: 20421597 Free PMC article.
-
Telomerase-null survivor screening identifies novel telomere recombination regulators.PLoS Genet. 2013;9(1):e1003208. doi: 10.1371/journal.pgen.1003208. Epub 2013 Jan 17. PLoS Genet. 2013. PMID: 23390378 Free PMC article.
-
Recombination-based telomere maintenance is dependent on Tel1-MRN and Rap1 and inhibited by telomerase, Taz1, and Ku in fission yeast.Mol Cell Biol. 2008 Mar;28(5):1443-55. doi: 10.1128/MCB.01614-07. Epub 2007 Dec 26. Mol Cell Biol. 2008. PMID: 18160711 Free PMC article.
-
Recombination can either help maintain very short telomeres or generate longer telomeres in yeast cells with weak telomerase activity.Eukaryot Cell. 2011 Aug;10(8):1131-42. doi: 10.1128/EC.05079-11. Epub 2011 Jun 10. Eukaryot Cell. 2011. PMID: 21666075 Free PMC article.
References
-
- Artandi, S. E., and R. A. DePinho. 2000. A critical role for telomeres in suppressing and facilitating carcinogenesis. Curr. Opin. Genet. Dev. 10:39-46. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
