A functional screen for Myc-responsive genes reveals serine hydroxymethyltransferase, a major source of the one-carbon unit for cell metabolism
- PMID: 12138190
- PMCID: PMC133987
- DOI: 10.1128/MCB.22.16.5793-5800.2002
A functional screen for Myc-responsive genes reveals serine hydroxymethyltransferase, a major source of the one-carbon unit for cell metabolism
Abstract
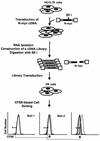
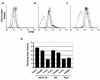
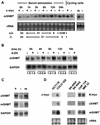
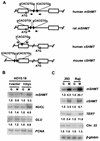
A cDNA library enriched with Myc-responsive cDNAs but depleted of myc cDNAs was used in a functional screen for growth enhancement in c-myc-null cells. A cDNA clone for mitochondrial serine hydroxymethyltransferase (mSHMT) that was capable of partial complementation of the growth defects of c-myc-null cells was identified. Expression analysis and chromatin immunoprecipitation demonstrated that mSHMT is a direct Myc target gene. Furthermore, a separate gene encoding the cytoplasmic isoform of the same enzyme is also a direct target of Myc regulation. SHMT enzymes are the major source of the one-carbon unit required for folate metabolism and for the biosynthesis of nucleotides and amino acids. Our data establish a novel functional link between Myc and the regulation of cellular metabolism.
Figures
Similar articles
-
Molecular cloning, characterization, and regulation of the human mitochondrial serine hydroxymethyltransferase gene.J Biol Chem. 1997 Jan 17;272(3):1842-8. doi: 10.1074/jbc.272.3.1842. J Biol Chem. 1997. PMID: 8999870
-
Inactivation of the cytosolic and mitochondrial serine hydroxymethyl transferase genes in Leishmania major.Mol Biochem Parasitol. 2015 Dec;204(2):106-110. doi: 10.1016/j.molbiopara.2016.02.003. Epub 2016 Feb 8. Mol Biochem Parasitol. 2015. PMID: 26868981
-
Is mutated serine hydroxymethyltransferase (SHMT) involved in the etiology of neural tube defects?Mol Genet Metab. 2001 Jun;73(2):164-72. doi: 10.1006/mgme.2001.3175. Mol Genet Metab. 2001. PMID: 11386852
-
Serine hydroxymethyltransferase: a model enzyme for mechanistic, structural, and evolutionary studies.Biochim Biophys Acta. 2011 Nov;1814(11):1489-96. doi: 10.1016/j.bbapap.2010.10.010. Epub 2010 Nov 5. Biochim Biophys Acta. 2011. PMID: 21059411 Review.
-
Genetic manipulation of glycine decarboxylation.J Exp Bot. 2003 Jun;54(387):1523-35. doi: 10.1093/jxb/erg171. Epub 2003 Apr 28. J Exp Bot. 2003. PMID: 12730263 Review.
Cited by
-
Therapeutic Targeting of Mitochondrial One-Carbon Metabolism in Cancer.Mol Cancer Ther. 2020 Nov;19(11):2245-2255. doi: 10.1158/1535-7163.MCT-20-0423. Epub 2020 Sep 2. Mol Cancer Ther. 2020. PMID: 32879053 Free PMC article. Review.
-
Mouse genetics suggests cell-context dependency for Myc-regulated metabolic enzymes during tumorigenesis.PLoS Genet. 2012;8(3):e1002573. doi: 10.1371/journal.pgen.1002573. Epub 2012 Mar 15. PLoS Genet. 2012. PMID: 22438825 Free PMC article.
-
Targeting HOTAIRM1 ameliorates glioblastoma by disrupting mitochondrial oxidative phosphorylation and serine metabolism.iScience. 2022 Jul 31;25(8):104823. doi: 10.1016/j.isci.2022.104823. eCollection 2022 Aug 19. iScience. 2022. PMID: 35992092 Free PMC article.
-
Cysteine metabolic circuitries: druggable targets in cancer.Br J Cancer. 2021 Mar;124(5):862-879. doi: 10.1038/s41416-020-01156-1. Epub 2020 Nov 23. Br J Cancer. 2021. PMID: 33223534 Free PMC article. Review.
-
The Influence of Mitochondrial Energy and 1C Metabolism on the Efficacy of Anticancer Drugs: Exploring Potential Mechanisms of Resistance.Curr Med Chem. 2023;30(11):1209-1231. doi: 10.2174/0929867329666220401110418. Curr Med Chem. 2023. PMID: 35366764 Review.
References
-
- Berns, K., C. Martins, J. H. Dannenberg, A. Berns, H. te Riele, and R. Bernards. 2000. A genetic screen to identify genes that rescue the slow growth phenotype of c-myc null fibroblasts. Oncogene 19:3330-3334. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
