Site-directed perturbation of protein kinase C- integrin interaction blocks carcinoma cell chemotaxis
- PMID: 12138200
- PMCID: PMC133968
- DOI: 10.1128/MCB.22.16.5897-5911.2002
Site-directed perturbation of protein kinase C- integrin interaction blocks carcinoma cell chemotaxis
Abstract
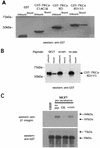
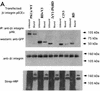
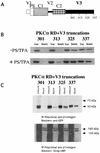
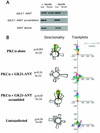
Polarized cell movement is an essential requisite for cancer metastasis; thus, interference with the tumor cell motility machinery would significantly modify its metastatic behavior. Protein kinase C alpha (PKC alpha) has been implicated in the promotion of a migratory cell phenotype. We report that the phorbol ester-induced cell polarization and directional motility in breast carcinoma cells is determined by a 12-amino-acid motif (amino acids 313 to 325) within the PKC alpha V3 hinge domain. This motif is also required for a direct association between PKC alpha and beta 1 integrin. Efficient binding of beta 1 integrin to PKC alpha requires the presence of both NPXY motifs (Cyto-2 and Cyto-3) in the integrin distal cytoplasmic domains. A cell-permeant inhibitor based on the PKC-binding sequence of beta 1 integrin was shown to block both PKC alpha-driven and epidermal growth factor (EGF)-induced chemotaxis. When introduced as a minigene by retroviral transduction into human breast carcinoma cells, this inhibitor caused a striking reduction in chemotaxis towards an EGF gradient. Taken together, these findings identify a direct link between PKC alpha and beta 1 integrin that is critical for directed tumor cell migration. Importantly, our findings outline a new concept as to how carcinoma cell chemotaxis is enhanced and provide a conceptual basis for interfering with tumor cell dissemination.
Figures









References
-
- Bastiaens, P. I. H., and T. M. Jovin. 1998. Fluorescence resonance energy transfer microscopy, p. 136-146. In J. Celis (ed.), Cell biology: a laboratory handbook. Academic Press, New York, N.Y.
-
- Berditchevski, F., G. Bazzoni, and M. E. Hemler. 1995. Specific association of CD63 with the VLA-3 and VLA-6 integrins. J. Biol. Chem. 270:17784-17790. - PubMed
-
- Bretscher, M. S. 1996. Moving membrane up to the front of migrating cells. Cell 85:465-467. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
