R-Ras3/M-Ras induces neuronal differentiation of PC12 cells through cell-type-specific activation of the mitogen-activated protein kinase cascade
- PMID: 12138204
- PMCID: PMC133986
- DOI: 10.1128/MCB.22.16.5946-5961.2002
R-Ras3/M-Ras induces neuronal differentiation of PC12 cells through cell-type-specific activation of the mitogen-activated protein kinase cascade
Abstract
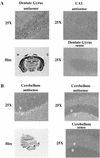
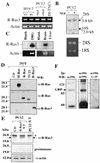
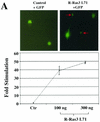
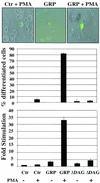
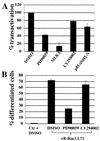
R-Ras3/M-Ras is a novel member of the Ras subfamily of GTP-binding proteins which has a unique expression pattern highly restricted to the mammalian central nervous system. In situ hybridization using an R-Ras3 cRNA probe revealed high levels of R-Ras3 transcripts in the hippocampal region of the mouse brain as well as a pattern of expression in the cerebellum that was distinct from that of H-Ras. We found that R-Ras3 was activated by nerve growth factor (NGF) and basic fibroblast growth factor as well as by the guanine nucleotide exchange factor GRP but not by epidermal growth factor. Ectopic expression of either R-Ras3 or GRP in PC12 cells induced efficient neuronal differentiation. The ability of NGF as well as GRP to promote differentiation of PC12 cells was attenuated by an R-Ras3 dominant-negative mutant. Furthermore, the biological action of R-Ras3 in PC12 cells was dependent on the mitogen-activated protein kinase (MAPK). Interestingly, whereas R-Ras3 was unable to mediate efficient activation of MAPK activity in NIH 3T3 cells, it was able to do so in PC12 cells. This cell-type specificity is in stark contrast to that of H-Ras, which can stimulate the MAPK pathway in both cell types. Indeed, this pattern of MAPK activation could be explained by the fact that R-Ras3 was unable to activate c-Raf, while it bound and stimulated the neuronal Raf isoform, B-Raf, in PC12 cells. Thus, R-Ras3 is implicated in a novel pathway of neuronal differentiation by coupling specific trophic factors to the MAPK cascade through the activation of B-Raf.
Figures






Similar articles
-
R-Ras3, a brain-specific Ras-related protein, activates Akt and promotes cell survival in PC12 cells.Oncogene. 2000 Apr 13;19(16):2014-22. doi: 10.1038/sj.onc.1203530. Oncogene. 2000. PMID: 10803462
-
Rit contributes to nerve growth factor-induced neuronal differentiation via activation of B-Raf-extracellular signal-regulated kinase and p38 mitogen-activated protein kinase cascades.Mol Cell Biol. 2005 Jan;25(2):830-46. doi: 10.1128/MCB.25.2.830-846.2005. Mol Cell Biol. 2005. PMID: 15632082 Free PMC article.
-
Mapping of atypical protein kinase C within the nerve growth factor signaling cascade: relationship to differentiation and survival of PC12 cells.Mol Cell Biol. 2000 Jul;20(13):4494-504. doi: 10.1128/MCB.20.13.4494-4504.2000. Mol Cell Biol. 2000. PMID: 10848576 Free PMC article.
-
The insulin/Ras pathway of adipocytic differentiation of 3T3 L1 cells: dissociation between Raf-1 kinase and the MAPK/RSK cascade.Int J Obes Relat Metab Disord. 1996 Mar;20 Suppl 3:S43-51. Int J Obes Relat Metab Disord. 1996. PMID: 8680477 Review.
-
Rit subfamily small GTPases: regulators in neuronal differentiation and survival.Cell Signal. 2013 Oct;25(10):2060-8. doi: 10.1016/j.cellsig.2013.06.002. Epub 2013 Jun 11. Cell Signal. 2013. PMID: 23770287 Free PMC article. Review.
Cited by
-
Autism-associated PTEN missense mutation leads to enhanced nuclear localization and neurite outgrowth in an induced pluripotent stem cell line.FEBS J. 2020 Nov;287(22):4848-4861. doi: 10.1111/febs.15287. Epub 2020 Mar 26. FEBS J. 2020. PMID: 32150788 Free PMC article.
-
Gene function in early mouse embryonic stem cell differentiation.BMC Genomics. 2007 Mar 29;8:85. doi: 10.1186/1471-2164-8-85. BMC Genomics. 2007. PMID: 17394647 Free PMC article.
-
R-Ras1 and R-Ras2 Expression in Anatomical Regions and Cell Types of the Central Nervous System.Int J Mol Sci. 2022 Jan 17;23(2):978. doi: 10.3390/ijms23020978. Int J Mol Sci. 2022. PMID: 35055164 Free PMC article.
-
R-Ras1 and R-Ras2 Are Essential for Oligodendrocyte Differentiation and Survival for Correct Myelination in the Central Nervous System.J Neurosci. 2018 May 30;38(22):5096-5110. doi: 10.1523/JNEUROSCI.3364-17.2018. Epub 2018 May 2. J Neurosci. 2018. PMID: 29720552 Free PMC article.
-
DA-Raf1, a competent intrinsic dominant-negative antagonist of the Ras-ERK pathway, is required for myogenic differentiation.J Cell Biol. 2007 Jun 4;177(5):781-93. doi: 10.1083/jcb.200703195. Epub 2007 May 29. J Cell Biol. 2007. PMID: 17535970 Free PMC article.
References
-
- Barbacid, M. 1987. ras genes. Annu. Rev. Biochem. 56:779-827. - PubMed
-
- Bar-Sagi, D., and J. R. Feramisco. 1985. Microinjection of the ras oncogene protein into PC12 cells induces morphological differentiation. Cell 42:841-848. - PubMed
-
- Boguski, M. S., and F. McCormick. 1993. Proteins regulating Ras and its relatives. Nature 366:643-654. - PubMed
-
- Bos, J. L. 1989. ras oncogenes in human cancer: a review. Cancer Res. 49:4682-4689. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
