Cell cycle arrest and repression of cyclin D1 transcription by INI1/hSNF5
- PMID: 12138206
- PMCID: PMC133966
- DOI: 10.1128/MCB.22.16.5975-5988.2002
Cell cycle arrest and repression of cyclin D1 transcription by INI1/hSNF5
Abstract
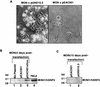
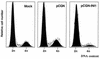
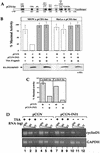
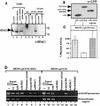
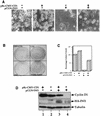
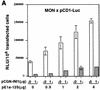
INI1/hSNF5 is a component of the ATP-dependent chromatin remodeling hSWI/SNF complex and a tumor suppressor gene of aggressive pediatric atypical teratoid and malignant rhabdoid tumors (AT/RT). To understand the molecular mechanisms underlying its tumor suppressor function, we studied the effect of reintroduction of INI1/hSNF5 into AT/RT-derived cell lines such as MON that carry biallelic deletions of the INI1/hSNF5 locus. We demonstrate that expression of INI1/hSNF5 causes G(0)-G(1) arrest and flat cell formation in these cells. In addition, INI1/hSNF5 repressed transcription of cyclin D1 gene in MON, in a histone deacetylase (HDAC)-dependent manner. Chromatin immunoprecipitation studies revealed that INI1/hSNF5 was directly recruited to the cyclin D1 promoter and that its binding correlated with recruitment of HDAC1 and deacetylation of histones at the promoter. Analysis of INI1/hSNF5 truncations indicated that cyclin D1 repression and flat cell formation are tightly correlated. Coexpression of cyclin D1 from a heterologous promoter in MON was sufficient to eliminate the INI1-mediated flat cell formation and cell cycle arrest. Furthermore, cyclin D1 was overexpressed in AT/RT tumors. Our data suggest that one of the mechanisms by which INI1/hSNF5 exerts its tumor suppressor function is by mediating the cell cycle arrest due to the direct recruitment of HDAC activity to the cyclin D1 promoter thereby causing its repression and G(0)-G(1) arrest. Repression of cyclin D1 gene expression may serve as a useful strategy to treat AT/RT.
Figures


Similar articles
-
Aurora A is a repressed effector target of the chromatin remodeling protein INI1/hSNF5 required for rhabdoid tumor cell survival.Cancer Res. 2011 May 1;71(9):3225-35. doi: 10.1158/0008-5472.CAN-10-2167. Epub 2011 Apr 26. Cancer Res. 2011. PMID: 21521802
-
A key role of the hSNF5/INI1 tumour suppressor in the control of the G1-S transition of the cell cycle.Oncogene. 2002 Sep 19;21(42):6403-12. doi: 10.1038/sj.onc.1205841. Oncogene. 2002. PMID: 12226744
-
Cyclin D1 is overexpressed in atypical teratoid/rhabdoid tumor with hSNF5/INI1 gene inactivation.J Neurooncol. 2005 Jun;73(2):117-24. doi: 10.1007/s11060-004-4276-4. J Neurooncol. 2005. PMID: 15981100
-
Integrase interactor 1 in health and disease.Curr Protein Pept Sci. 2015;16(6):478-90. doi: 10.2174/1389203716666150316115156. Curr Protein Pept Sci. 2015. PMID: 25772157 Review.
-
Mechanisms by which SMARCB1 loss drives rhabdoid tumor growth.Cancer Genet. 2014 Sep;207(9):365-72. doi: 10.1016/j.cancergen.2014.04.004. Epub 2014 Apr 13. Cancer Genet. 2014. PMID: 24853101 Free PMC article. Review.
Cited by
-
The histone deacetylase inhibitor SAHA acts in synergism with fenretinide and doxorubicin to control growth of rhabdoid tumor cells.BMC Cancer. 2013 Jun 13;13:286. doi: 10.1186/1471-2407-13-286. BMC Cancer. 2013. PMID: 23764045 Free PMC article.
-
Epigenetic dysregulation in cancer.Am J Pathol. 2009 Oct;175(4):1353-61. doi: 10.2353/ajpath.2009.081142. Epub 2009 Aug 28. Am J Pathol. 2009. PMID: 19717641 Free PMC article.
-
SWI/SNF nucleosome remodellers and cancer.Nat Rev Cancer. 2011 Jun 9;11(7):481-92. doi: 10.1038/nrc3068. Nat Rev Cancer. 2011. PMID: 21654818 Review.
-
Generation of a mouse model of atypical teratoid/rhabdoid tumor of the central nervous system through combined deletion of Snf5 and p53.Cancer Res. 2015 Nov 1;75(21):4629-39. doi: 10.1158/0008-5472.CAN-15-0874. Epub 2015 Sep 11. Cancer Res. 2015. PMID: 26363008 Free PMC article.
-
BRG1 controls the activity of the retinoblastoma protein via regulation of p21CIP1/WAF1/SDI.Mol Cell Biol. 2004 Feb;24(3):1188-99. doi: 10.1128/MCB.24.3.1188-1199.2004. Mol Cell Biol. 2004. PMID: 14729964 Free PMC article.
References
-
- Albanese, C., M. D'Amico, A. T. Reutens, M. Fu, G. Watanabe, R. J. Lee, R. N. Kitsis, B. Henglein, M. Avantaggiati, K. Somasundarum, B. Thimmapaya, and R. G. Pestell. 1999. Activation of the cyclinD1 gene by the E1a associated protein p300 through AP-1 inhibits cellular apoptosis. J. Biol. Chem. 274:34186-34195. - PubMed
-
- Biegel, J. A., C. S. Allen, K. Kawasaki, N. Shimizu, M. L. Budarf, and C. J. Bell. 1996. Narrowing the critical region for a rhabdoid tumor locus in 22q11. Genes Chromosomes Cancer 16:94-105. - PubMed
-
- Biegel, J. A., J. Y. Zhou, L. B. Rorke, C. Stenstrom, L. M. Wainwright, and B. Fogelgren. 1999. Germ-line and acquired mutations of INI1 in atypical teratoid and rhabdoid tumors. Cancer Res. 59:74-79. - PubMed
-
- Carlson, M., and B. C. Laurent. 1994. The SNF/SWI family of global transcriptional activators. Curr. Opin. Cell Biol. 6:396-402. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
