Increased telomere length and hypersensitivity to DNA damaging agents in an Arabidopsis KU70 mutant
- PMID: 12140324
- PMCID: PMC137071
- DOI: 10.1093/nar/gkf445
Increased telomere length and hypersensitivity to DNA damaging agents in an Arabidopsis KU70 mutant
Abstract
We have identified a putative homologue of the KU70 gene (AtKU70) from Arabidopsis thaliana. In order to study its function in plants we have isolated an A.thaliana line that contains a T-DNA inserted into AtKU70. Plants homozygous for this insertion appear normal and are fertile. In other organisms the KU70 gene has been shown to play a role in the repair of DNA damage induced by ionising radiation (IR) and by radiomimetic chemicals such as methylmethane sulfonate (MMS). We show that AtKU70(-/-) plants are hypersensitive to IR and MMS, and thus the AtKU70 gene plays a similar role in DNA repair in plants as in other organisms. The KU70 gene also plays a role in maintaining telomere length. Yeast and mammalian cells deficient for Ku70 have shortened telomeres. When we studied the telomeres in the AtKU70(-/-) plants we found unexpectedly that they were significantly longer (>30 kb) than was found in wild-type plants (2-4 kb). We propose several hypotheses to explain this telomere lengthening in the AtKU70(-/-) plants.
Figures

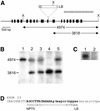

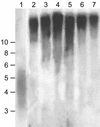
References
-
- Haber J.E. (2000) Recombination: a frank view of exchanges and vice versa. Curr. Opin. Cell Biol., 12, 286–292. - PubMed
-
- Kegel A., Sjostrand,J.O. and Astrom,S.U. (2001) Nej1p, a cell type-specific regulator of nonhomologous end joining in yeast. Curr. Biol., 11, 1611–1617. - PubMed
-
- Doherty A.J., Jackson,S.P. and Weller,G.R. (2001) Identification of bacterial homologues of the Ku DNA repair protein. FEBS Lett., 500, 186–188. - PubMed
-
- Featherstone C. and Jackson,S.P. (1999) Ku, a DNA repair protein with multiple cellular functions? Mutat. Res., 434, 3–15. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

