Identification of 5-formyluracil DNA glycosylase activity of human hNTH1 protein
- PMID: 12140329
- PMCID: PMC137084
- DOI: 10.1093/nar/gkf460
Identification of 5-formyluracil DNA glycosylase activity of human hNTH1 protein
Abstract
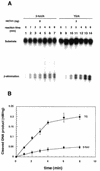
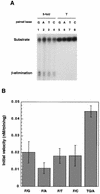
5-formyluracil (5-foU) is a potentially mutagenic lesion of thymine produced in DNA by ionizing radiation and various chemical oxidants. The elucidation of repair mechanisms for 5-foU will yield important insights into the biological consequences of the lesion. Recently, we reported that 5-foU is recognized and removed from DNA by Escherichia coli enzymes Nth (endonuclease III), Nei (endonuclease VIII) and MutM (formamidopyrimidine DNA glycosylase). Human cells have been shown to have enzymatic activities that release 5-foU from X-ray-irradiated DNA, but the molecular identities of these activities are not yet known. In this study, we demonstrate that human hNTH1 (endonuclease III homolog) has a DNA glycosylase/AP lyase activity that recognizes 5-foU in DNA and removes it. hNTH1 cleaved 5-foU-containing duplex oligonucleotides via a beta-elimination reaction. It formed Schiff base intermediates with 5-foU-containing oligonucleotides. Furthermore, hNTH1 cleaved duplex oligonucleotides containing all of the 5-foU/N pairs (N = G, A, T or C). The specific activities of hNTH1 for cleavage of oligonucleotides containing 5-foU and thymine glycol were 0.011 and 0.045 nM/min/ng protein, respectively. These results indicate that hNTH1 has DNA glycosylase activity with the potential to recognize 5-foU in DNA and remove it in human cells.
Figures



References
-
- Halliwell B. and Gutteridge,J.M.C. (1990) Role of free radicals and catalytic metal ions in human disease: overview. Methods Enzymol., 186, 1–85. - PubMed
-
- Wallace S.S. (1997) Oxidative damage to DNA and its repair. In Scandalios,J.G. (ed.), Oxidative Stress and the Molecular Biology of Antioxidant Defenses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 49–81.
-
- Cadet J., Delatour,T., Douki,T., Gasparutto,D., Pouget,J.P., Ravanat,J.L. and Sauvaigo,S. (1999) Hydroxyl radicals and DNA base damage. Mutat. Res., 424, 9–21. - PubMed
-
- Friedberg E.C., Walker,G. and Siede,W. (1995) DNA Repair and Mutagenesis. ASM Press, Washington, DC.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

