PRIMROSE: a computer program for generating and estimating the phylogenetic range of 16S rRNA oligonucleotide probes and primers in conjunction with the RDP-II database
- PMID: 12140334
- PMCID: PMC137075
- DOI: 10.1093/nar/gkf450
PRIMROSE: a computer program for generating and estimating the phylogenetic range of 16S rRNA oligonucleotide probes and primers in conjunction with the RDP-II database
Abstract
We describe PRIMROSE, a computer program for identifying 16S rRNA probes and PCR primers for use as phylogenetic and ecological tools in the identification and enumeration of bacteria. PRIMROSE is designed to use data from the Ribosomal Database Project (RDP) to find potentially useful oligonucleotides with up to two degenerate positions. The taxonomic range of these, and other existing oligonucleotides, can then be explored, allowing for the rapid identification of suitable oligonucleotides. PRIMROSE includes features to allow user-defined sequence databases to be used. An in silico trial of the program using the RDP database identified oligonucleotides that described their target taxa with a degree of accuracy far greater than that of equivalent currently used oligonucleotides. We identify oligonucleotides for subdivisions of the Proteobacteria and for the Cytophaga-Flexibacter-Bacteroides (CFB) division. These oligonucleotides describe up to 94.7% of their target taxon with fewer than 50 non-target hits, and the authors recommend that they be investigated further. A comparison with PROBE DESIGN within the ARB software package shows that PRIMROSE is capable of identifying oligonucleotides with a higher specificity. PRIMROSE has an intuitive graphical user interface and runs on the Microsoft Windows 95/NT/2000 operating systems. It is open source and is freely available from the authors.
Figures


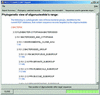
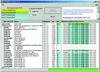
References
-
- Amann R. and Ludwig,W. (2000) Ribosomal RNA-targeted nucleic acid probes for studies in microbial ecology. FEMS Microbiol. Rev., 24, 555–565. - PubMed
-
- Stahl D.A. and Amann,R. (1991) Development and application of nucleic acid probes. In Stackebrandt,E. and Goodfellow,M. (eds), Nucleic Acid Techniques in Bacterial Systematics. Wiley-Interscience, Chichester, UK, pp. 205–248.
-
- Hyndman D., Cooper,A., Pruzinsky,S., Coad,D. and Mitsuhashi,M. (1996) Software to determine optimal oligonucleotide sequences based on hybridization simulation data. Biotechniques, 20, 1090–1097. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

