Comparison of genome structures of vibrios, bacteria possessing two chromosomes
- PMID: 12142404
- PMCID: PMC135242
- DOI: 10.1128/JB.184.16.4351-4358.2002
Comparison of genome structures of vibrios, bacteria possessing two chromosomes
Abstract
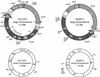
Vibrios are gram-negative gamma-proteobacteria which are ubiquitous in marine and estuarine environments. Recently, we demonstrated that some, if not all, Vibrio species have two circular chromosomes. The whole genome sequence of Vibrio cholerae N16961 has been reported. In this study, we constructed a physical and genetic map of the genome of Kanagawa phenomenon-positive Vibrio parahaemolyticus strain KX-V237 and compared it with those of V. parahaemolyticus AQ4673 and V. cholerae N16961. The genome of KX-V237 comprised two circular chromosomes (3.3 and 1.9 Mb), similar to the structure of the AQ4673 genome. The relative positions of the genes on the genomes were well conserved in the two strains, but a large inversion on the large chromosomes, probably symmetric around the replication origin, was suggested. Although the sizes of the large chromosomes of KX-V237 and V. cholerae N16961 were similar, the sizes of the small chromosomes were very different. Unlike N16961, the superintegron of KX-V237 was located on the large chromosome. Comparison of the genetic maps of the chromosomes of KX-V237 and V. cholerae N16961 revealed that most of the open reading frames (ORFs) present on the large chromosome of the V. cholerae strain had homologues on the large chromosome of the V. parahaemolyticus strain and that most of the ORFs on the small chromosome of N16961 were present on the small chromosome of KX-V237. The difference in the orders of the ORFs on the chromosomes of N16961 and KX-V237 implies that numerous and frequent genetic exchanges have occurred intrachromosomally rather than interchromosomally.
Figures



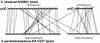
Similar articles
-
Physical and genetic map of the genome of Vibrio parahaemolyticus: presence of two chromosomes in Vibrio species.Mol Microbiol. 1999 Mar;31(5):1513-21. doi: 10.1046/j.1365-2958.1999.01296.x. Mol Microbiol. 1999. PMID: 10200969
-
[Vibrios (Vibrio cholerae, V. parahaemolyticus, V. vulnificus)].Nihon Rinsho. 2003 Mar;61 Suppl 3:722-6. Nihon Rinsho. 2003. PMID: 12718054 Review. Japanese. No abstract available.
-
Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V cholerae.Lancet. 2003 Mar 1;361(9359):743-9. doi: 10.1016/S0140-6736(03)12659-1. Lancet. 2003. PMID: 12620739
-
In silico comparative study of the genomic islands of Vibrio cholerae MJ1236 with those of Classical and El Tor N16961 strains of Vibrio cholerae.FEMS Microbiol Lett. 2011 Aug;321(1):75-81. doi: 10.1111/j.1574-6968.2011.02316.x. Epub 2011 Jun 6. FEMS Microbiol Lett. 2011. PMID: 21599729
-
Management of multipartite genomes: the Vibrio cholerae model.Curr Opin Microbiol. 2014 Dec;22:120-6. doi: 10.1016/j.mib.2014.10.003. Curr Opin Microbiol. 2014. PMID: 25460805 Review.
Cited by
-
Core and Accessory Genome Analysis of Vibrio mimicus.Microorganisms. 2021 Jan 18;9(1):191. doi: 10.3390/microorganisms9010191. Microorganisms. 2021. PMID: 33477474 Free PMC article.
-
Simple sequence repeats and compositional bias in the bipartite Ralstonia solanacearum GMI1000 genome.BMC Genomics. 2003 Mar 17;4(1):10. doi: 10.1186/1471-2164-4-10. Epub 2003 Mar 17. BMC Genomics. 2003. PMID: 12697060 Free PMC article.
-
The Pelagic Bacterium Paraphotobacterium marinum Has the Smallest Complete Genome Within the Family Vibrionaceae.Front Microbiol. 2017 Oct 11;8:1994. doi: 10.3389/fmicb.2017.01994. eCollection 2017. Front Microbiol. 2017. PMID: 29085348 Free PMC article.
-
Toxin-linked mobile genetic elements in major enteric bacterial pathogens.Gut Microbiome (Camb). 2023 Mar 17;4:e5. doi: 10.1017/gmb.2023.2. eCollection 2023. Gut Microbiome (Camb). 2023. PMID: 39295911 Free PMC article. Review.
-
Determination of the transcriptome of Vibrio cholerae during intraintestinal growth and midexponential phase in vitro.Proc Natl Acad Sci U S A. 2003 Feb 4;100(3):1286-91. doi: 10.1073/pnas.0337479100. Epub 2003 Jan 27. Proc Natl Acad Sci U S A. 2003. PMID: 12552086 Free PMC article.
References
-
- Centers for Disease Control and Prevention. 1998. Outbreak of Vibrio parahaemolyticus infections associated with eating raw oysters—Pacific Northwest, 1997. Morb. Mortal. Wkly. Rep. 47:457-462. - PubMed
-
- Centers for Disease Control and Prevention. 1999. Outbreak of Vibrio parahaemolyticus infection associated with eating raw oysters and clams harvested from Long Island Sound—Connecticut, New Jersey, and New York, 1998. Morb. Mortal. Wkly. Rep. 48:48-51. - PubMed
-
- Clark, C. A., L. Purins, P. Kaewrakon, and P. A. Manning. 1997. VCR repetitive sequence elements in the Vibrio cholerae chromosome constitute a mega-integron. Mol. Microbiol. 26:1137-1138. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

