Genes of de novo pyrimidine biosynthesis from the hyperthermoacidophilic crenarchaeote Sulfolobus acidocaldarius: novel organization in a bipolar operon
- PMID: 12142413
- PMCID: PMC135248
- DOI: 10.1128/JB.184.16.4430-4441.2002
Genes of de novo pyrimidine biosynthesis from the hyperthermoacidophilic crenarchaeote Sulfolobus acidocaldarius: novel organization in a bipolar operon
Abstract
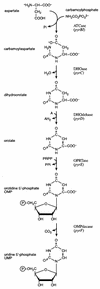
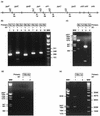
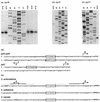
Sequencing a 8,519-bp segment of the Sulfolobus acidocaldarius genome revealed the existence of a tightly packed bipolar pyrimidine gene cluster encoding the enzymes of de novo UMP synthesis. The G+C content of 35.3% is comparable to that of the entire genome, but intergenic regions exhibit a considerably lower percentage of strong base pairs. Coding regions harbor the classical excess of purines on the coding strand, whereas intergenic regions do not show this bias. Reverse transcription-PCR and primer extension experiments demonstrated the existence of two polycistronic messengers, pyrEF-orf8 and pyrBI-orf1-pyrCD-orf2-orf3-orf4, initiated from a pair of divergent and partially overlapping promoters. The gene order and the grouping in two wings of a bipolar operon constitute a novel organization of pyr genes that also occurs in the recently determined genome sequences of Sulfolobus solfataricus P2 and Sulfolobus tokodaii strain 7; the configuration appears therefore characteristic of Sulfolobus. The quasi-leaderless pyrE and pyrB genes do not bear a Shine-Dalgarno sequence, whereas the initiation codon of promoter-distal genes is preceded at an appropriate distance by a sequence complementary to the 3' end of 16S rRNA. The polycistronic nature of the pyr messengers and the existence of numerous overlaps between contiguous open reading frames suggests the existence of translational coupling. pyrB transcription was shown to be approximately twofold repressed in the presence of uracil. The mechanism underlying this modulation is as yet unknown, but it appears to be of a type different from the various attenuation-like mechanisms that regulate pyrB transcription in bacteria. In contrast, the pyrE-pyrB promoter/control region harbors direct repeats and imperfect palindromes reminiscent of target sites for the binding of a hypothetical regulatory protein(s).
Figures
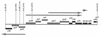

Similar articles
-
Functional organization and nucleotide sequence of the Bacillus subtilis pyrimidine biosynthetic operon.J Biol Chem. 1991 May 15;266(14):9113-27. J Biol Chem. 1991. PMID: 1709162
-
The pyrimidine biosynthesis operon of the thermophile Bacillus caldolyticus includes genes for uracil phosphoribosyltransferase and uracil permease.J Bacteriol. 1994 Jun;176(12):3698-707. doi: 10.1128/jb.176.12.3698-3707.1994. J Bacteriol. 1994. PMID: 8206848 Free PMC article.
-
Regulation of the Bacillus subtilis pyrimidine biosynthetic (pyr) gene cluster by an autogenous transcriptional attenuation mechanism.J Bacteriol. 1994 Jun;176(12):3708-22. doi: 10.1128/jb.176.12.3708-3722.1994. J Bacteriol. 1994. PMID: 8206849 Free PMC article.
-
Regulation of pyrimidine biosynthetic gene expression in bacteria: repression without repressors.Microbiol Mol Biol Rev. 2008 Jun;72(2):266-300, table of contents. doi: 10.1128/MMBR.00001-08. Microbiol Mol Biol Rev. 2008. PMID: 18535147 Free PMC article. Review.
-
Regulation of the Bacillus subtilis pyrimidine biosynthetic operon by transcriptional attenuation: control of gene expression by an mRNA-binding protein.Prog Nucleic Acid Res Mol Biol. 1999;62:329-67. doi: 10.1016/s0079-6603(08)60512-7. Prog Nucleic Acid Res Mol Biol. 1999. PMID: 9932459 Review.
Cited by
-
Molecular characteristics of spontaneous deletions in the hyperthermophilic archaeon Sulfolobus acidocaldarius.J Bacteriol. 2003 Feb;185(4):1266-72. doi: 10.1128/JB.185.4.1266-1272.2003. J Bacteriol. 2003. PMID: 12562797 Free PMC article.
-
Antiviral and Antimicrobial Nucleoside Derivatives: Structural Features and Mechanisms of Action.Mol Biol. 2021;55(6):786-812. doi: 10.1134/S0026893321040105. Epub 2021 Dec 17. Mol Biol. 2021. PMID: 34955556 Free PMC article.
-
Sa-Lrp from Sulfolobus acidocaldarius is a versatile, glutamine-responsive, and architectural transcriptional regulator.Microbiologyopen. 2013 Feb;2(1):75-93. doi: 10.1002/mbo3.58. Epub 2012 Dec 16. Microbiologyopen. 2013. PMID: 23255531 Free PMC article.
-
An active nonautonomous mobile element in Sulfolobus islandicus REN1H1.J Bacteriol. 2007 Mar;189(5):2145-9. doi: 10.1128/JB.01567-06. Epub 2006 Dec 8. J Bacteriol. 2007. PMID: 17158679 Free PMC article.
-
The genome of Sulfolobus acidocaldarius, a model organism of the Crenarchaeota.J Bacteriol. 2005 Jul;187(14):4992-9. doi: 10.1128/JB.187.14.4992-4999.2005. J Bacteriol. 2005. PMID: 15995215 Free PMC article.
References
-
- Brock, T. D., K. M. Brock, R. T. Belly, and R. L. Weiss. 1972. Sulfolobus: a new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch. Microbiol. 84:54-68. - PubMed
-
- Charlier, D., A. Kholti, N. Huysveld, D. Gigot, D. Maes, T.-L. Thia-Toong, and N. Glansdorff. 2000. Mutational analysis of Escherichia coli PepA, a multifunctional DNA-binding aminopeptidase. J. Mol. Biol. 302:411-426. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

