Suppressive subtractive hybridization detects extensive genomic diversity in Thermotoga maritima
- PMID: 12142418
- PMCID: PMC135253
- DOI: 10.1128/JB.184.16.4475-4488.2002
Suppressive subtractive hybridization detects extensive genomic diversity in Thermotoga maritima
Abstract
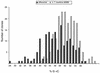
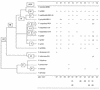
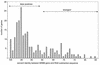
Comparisons between genomes of closely related bacteria often show large variations in gene content, even between strains of the same species. Such studies have focused mainly on pathogens; here, we examined Thermotoga maritima, a free-living hyperthermophilic bacterium, by using suppressive subtractive hybridization. The genome sequence of T. maritima MSB8 is available, and DNA from this strain served as a reference to obtain strain-specific sequences from Thermotoga sp. strain RQ2, a very close relative (approximately 96% identity for orthologous protein-coding genes, 99.7% identity in the small-subunit rRNA sequence). Four hundred twenty-six RQ2 subtractive clones were sequenced. One hundred sixty-six had no DNA match in the MSB8 genome. These differential clones comprise, in sum, 48 kb of RQ2-specific DNA and match 72 genes in the GenBank database. From the number of identical clones, we estimated that RQ2 contains 350 to 400 genes not found in MSB8. Assuming a similar genome size, this corresponds to 20% of the RQ2 genome. A large proportion of the RQ2-specific genes were predicted to be involved in sugar transport and polysaccharide degradation, suggesting that polysaccharides are more important as nutrients for this strain than for MSB8. Several clones encode proteins involved in the production of surface polysaccharides. RQ2 encodes multiple subunits of a V-type ATPase, while MSB8 possesses only an F-type ATPase. Moreover, an RQ2-specific MutS homolog was found among the subtractive clones and appears to belong to a third novel archaeal type MutS lineage. Southern blot analyses showed that some of the RQ2 differential sequences are found in some other members of the order Thermotogales, but the distribution of these variable genes is patchy, suggesting frequent lateral gene transfer within the group.
Figures


Similar articles
-
Evolution of mal ABC transporter operons in the Thermococcales and Thermotogales.BMC Evol Biol. 2008 Jan 15;8:7. doi: 10.1186/1471-2148-8-7. BMC Evol Biol. 2008. PMID: 18197971 Free PMC article.
-
Targeting clusters of transferred genes in Thermotoga maritima.Environ Microbiol. 2003 Nov;5(11):1144-54. doi: 10.1046/j.1462-2920.2003.00515.x. Environ Microbiol. 2003. PMID: 14641594
-
Genome sequence of Thermotoga sp. strain RQ2, a hyperthermophilic bacterium isolated from a geothermally heated region of the seafloor near Ribeira Quente, the Azores.J Bacteriol. 2011 Oct;193(20):5869-70. doi: 10.1128/JB.05923-11. J Bacteriol. 2011. PMID: 21952543 Free PMC article.
-
Microbial biochemistry, physiology, and biotechnology of hyperthermophilic Thermotoga species.FEMS Microbiol Rev. 2006 Nov;30(6):872-905. doi: 10.1111/j.1574-6976.2006.00039.x. FEMS Microbiol Rev. 2006. PMID: 17064285 Review.
-
Thermotoga heats up lateral gene transfer.Curr Biol. 1999 Oct 7;9(19):R747-51. doi: 10.1016/s0960-9822(99)80474-6. Curr Biol. 1999. PMID: 10531001 Review.
Cited by
-
Functional and comparative genomic analyses of an operon involved in fructooligosaccharide utilization by Lactobacillus acidophilus.Proc Natl Acad Sci U S A. 2003 Jul 22;100(15):8957-62. doi: 10.1073/pnas.1332765100. Epub 2003 Jul 7. Proc Natl Acad Sci U S A. 2003. PMID: 12847288 Free PMC article.
-
Several archaeal homologs of putative oligopeptide-binding proteins encoded by Thermotoga maritima bind sugars.Appl Environ Microbiol. 2006 Feb;72(2):1336-45. doi: 10.1128/AEM.72.2.1336-1345.2006. Appl Environ Microbiol. 2006. PMID: 16461685 Free PMC article.
-
PCR-based positive hybridization to detect genomic diversity associated with bacterial secondary metabolism.Nucleic Acids Res. 2004 Jan 12;32(1):e7. doi: 10.1093/nar/gnh012. Nucleic Acids Res. 2004. PMID: 14718552 Free PMC article.
-
Evolution of mal ABC transporter operons in the Thermococcales and Thermotogales.BMC Evol Biol. 2008 Jan 15;8:7. doi: 10.1186/1471-2148-8-7. BMC Evol Biol. 2008. PMID: 18197971 Free PMC article.
-
Divergence and redundancy of 16S rRNA sequences in genomes with multiple rrn operons.J Bacteriol. 2004 May;186(9):2629-35. doi: 10.1128/JB.186.9.2629-2635.2004. J Bacteriol. 2004. PMID: 15090503 Free PMC article.
References
-
- Alm, R. A., and T. J. Trust. 1999. Analysis of the genetic diversity of Helicobacter pylori: the tale of two genomes. J. Mol. Med. 77:834-846. - PubMed
-
- Beja, O., E. V. Koonin, L. Aravind, L. T. Taylor, H. Seitz, J. L. Stein, D. C. Bensen, R. A. Feldman, R. V. Swanson, and E. F. DeLong. 2002. Comparative genomic analysis of archaeal genotypic variants in a single population and in two different oceanic provinces. Appl. Environ. Microbiol. 68:335-345. - PMC - PubMed
-
- Bogush, M. L., T. V. Velikodvorskaya, Y. B. Lebedev, L. G. Nikolaev, S. A. Lukyanov, A. F. Fradkov, B. K. Pliyev, M. N. Boichenko, G. N. Usatova, A. A. Vorobiev, G. L. Anders, and E. D. Sverdlov. 1999. Identification and localization of differences between Escherichia coli and Salmonella typhimurium genomes by suppressive subtractive hybridization. Mol. Gen. Genet. 262:721-729. - PubMed
-
- Boucher, Y., C. L. Nesbø, and W. F. Doolittle. 2001. Microbial genomes: dealing with diversity. Curr. Opin. Microbiol. 4:285-289. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

