From genetic footprinting to antimicrobial drug targets: examples in cofactor biosynthetic pathways
- PMID: 12142426
- PMCID: PMC135229
- DOI: 10.1128/JB.184.16.4555-4572.2002
From genetic footprinting to antimicrobial drug targets: examples in cofactor biosynthetic pathways
Abstract
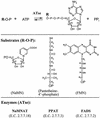
Novel drug targets are required in order to design new defenses against antibiotic-resistant pathogens. Comparative genomics provides new opportunities for finding optimal targets among previously unexplored cellular functions, based on an understanding of related biological processes in bacterial pathogens and their hosts. We describe an integrated approach to identification and prioritization of broad-spectrum drug targets. Our strategy is based on genetic footprinting in Escherichia coli followed by metabolic context analysis of essential gene orthologs in various species. Genes required for viability of E. coli in rich medium were identified on a whole-genome scale using the genetic footprinting technique. Potential target pathways were deduced from these data and compared with a panel of representative bacterial pathogens by using metabolic reconstructions from genomic data. Conserved and indispensable functions revealed by this analysis potentially represent broad-spectrum antibacterial targets. Further target prioritization involves comparison of the corresponding pathways and individual functions between pathogens and the human host. The most promising targets are validated by direct knockouts in model pathogens. The efficacy of this approach is illustrated using examples from metabolism of adenylate cofactors NAD(P), coenzyme A, and flavin adenine dinucleotide. Several drug targets within these pathways, including three distantly related adenylyltransferases (orthologs of the E. coli genes nadD, coaD, and ribF), are discussed in detail.
Figures

References
-
- Alm, R. A., L. S. L. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trost. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. - PubMed
-
- Arigoni, F., F. Talabot, M. Peitsch, M. D. Edgerton, E. Meldrum, E. Allet, R. Fish, T. Jamotte, M. L. Curchod, and H. Loferer. 1998. A genome-based approach for the identification of essential bacterial genes. Nat. Biotechnol. 16:851-856. - PubMed
-
- Bacher, A., S. Eberhardt, W. Eisenreich, M. Fischer, S. Herz, B. Illarionov, K. Kis, and G. Richter. 2001. Biosynthesis of riboflavin. Vitam. Horm. 61:2-49. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

