Genome-scale metabolic model of Helicobacter pylori 26695
- PMID: 12142428
- PMCID: PMC135230
- DOI: 10.1128/JB.184.16.4582-4593.2002
Genome-scale metabolic model of Helicobacter pylori 26695
Abstract
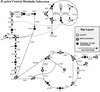
A genome-scale metabolic model of Helicobacter pylori 26695 was constructed from genome sequence annotation, biochemical, and physiological data. This represents an in silico model largely derived from genomic information for an organism for which there is substantially less biochemical information available relative to previously modeled organisms such as Escherichia coli. The reconstructed metabolic network contains 388 enzymatic and transport reactions and accounts for 291 open reading frames. Within the paradigm of constraint-based modeling, extreme-pathway analysis and flux balance analysis were used to explore the metabolic capabilities of the in silico model. General network properties were analyzed and compared to similar results previously generated for Haemophilus influenzae. A minimal medium required by the model to generate required biomass constituents was calculated, indicating the requirement of eight amino acids, six of which correspond to essential human amino acids. In addition a list of potential substrates capable of fulfilling the bulk carbon requirements of H. pylori were identified. A deletion study was performed wherein reactions and associated genes in central metabolism were deleted and their effects were simulated under a variety of substrate availability conditions, yielding a number of reactions that are deemed essential. Deletion results were compared to recently published in vitro essentiality determinations for 17 genes. The in silico model accurately predicted 10 of 17 deletion cases, with partial support for additional cases. Collectively, the results presented herein suggest an effective strategy of combining in silico modeling with experimental technologies to enhance biological discovery for less characterized organisms and their genomes.
Figures


References
-
- Arigoni, F., F. Talabot, M. Peitsch, M. D. Edgerton, E. Meldrum, E. Allet, R. Fish, T. Jamotte, M. L. Curchod, and H. Loferer. 1998. A genome-based approach for the identification of essential bacterial genes. Nat. Biotechnol. 16:851-856. - PubMed
-
- Birkholz, S., U. Knipp, E. Lemma, A. Kroger, and W. Opferkuch. 1994. Fumarate reductase of Helicobacter pylori—an immunogenic protein. J. Med. Microbiol. 41:56-62. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

