Functional annotation of class I lysyl-tRNA synthetase phylogeny indicates a limited role for gene transfer
- PMID: 12142429
- PMCID: PMC135231
- DOI: 10.1128/JB.184.16.4594-4600.2002
Functional annotation of class I lysyl-tRNA synthetase phylogeny indicates a limited role for gene transfer
Abstract
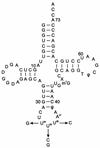
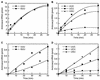
Functional and comparative genomic studies have previously shown that the essential protein lysyl-tRNA synthetase (LysRS) exists in two unrelated forms. Most prokaryotes and all eukaryotes contain a class II LysRS, whereas most archaea and a few bacteria contain a less common class I LysRS. In bacteria the class I LysRS is only found in the alpha-proteobacteria and a scattering of other groups, including the spirochetes, while the class I protein is by far the most common form of LysRS in archaea. To investigate this unusual distribution we functionally annotated a representative phylogenetic sampling of LysRS proteins. Class I LysRS proteins from a variety of bacteria and archaea were characterized in vitro by their ability to recognize Escherichia coli tRNA(Lys) anticodon mutants. Class I LysRS proteins were found to fall into two distinct groups, those that preferentially recognize the third anticodon nucleotide of tRNA(Lys) (U36) and those that recognize both the second and third positions (U35 and U36). Strong recognition of U35 and U36 was confined to the pyrococcus-spirochete grouping within the archaeal branch of the class I LysRS phylogenetic tree, while U36 recognition was seen in other archaea and an example from the alpha-proteobacteria. Together with the corresponding phylogenetic relationships, these results suggest that despite its comparative rarity the distribution of class I LysRS conforms to the canonical archaeal-bacterial division. The only exception, suggested from both functional and phylogenetic data, appears to be the horizontal transfer of class I LysRS from a pyrococcal progenitor to a limited number of bacteria.
Figures



Similar articles
-
Context-dependent anticodon recognition by class I lysyl-tRNA synthetases.Proc Natl Acad Sci U S A. 2000 Dec 19;97(26):14224-8. doi: 10.1073/pnas.97.26.14224. Proc Natl Acad Sci U S A. 2000. PMID: 11121028 Free PMC article.
-
Substrate recognition by class I lysyl-tRNA synthetases: a molecular basis for gene displacement.Proc Natl Acad Sci U S A. 1999 Jan 19;96(2):418-23. doi: 10.1073/pnas.96.2.418. Proc Natl Acad Sci U S A. 1999. PMID: 9892648 Free PMC article.
-
tRNA anticodon recognition and specification within subclass IIb aminoacyl-tRNA synthetases.J Mol Biol. 1998 May 15;278(4):801-13. doi: 10.1006/jmbi.1998.1711. J Mol Biol. 1998. PMID: 9614943
-
Lysyl-tRNA synthetase.Biol Chem Hoppe Seyler. 1995 Aug;376(8):451-72. doi: 10.1515/bchm3.1995.376.8.451. Biol Chem Hoppe Seyler. 1995. PMID: 7576245 Review.
-
Non-canonical roles of lysyl-tRNA synthetase in health and disease.Trends Mol Med. 2013 Dec;19(12):726-31. doi: 10.1016/j.molmed.2013.07.011. Epub 2013 Aug 20. Trends Mol Med. 2013. PMID: 23972532 Review.
Cited by
-
Human lysyl-tRNA synthetase is secreted to trigger proinflammatory response.Proc Natl Acad Sci U S A. 2005 May 3;102(18):6356-61. doi: 10.1073/pnas.0500226102. Epub 2005 Apr 25. Proc Natl Acad Sci U S A. 2005. PMID: 15851690 Free PMC article.
-
Recognition of pyrrolysine tRNA by the Desulfitobacterium hafniense pyrrolysyl-tRNA synthetase.Nucleic Acids Res. 2007;35(4):1270-8. doi: 10.1093/nar/gkl1151. Epub 2007 Jan 31. Nucleic Acids Res. 2007. PMID: 17267409 Free PMC article.
-
Structural Computational Analysis of the Natural History of Class I aminoacyl-tRNA Synthetases Suggests their Role in Establishing the Genetic Code.J Mol Evol. 2021 Dec;89(9-10):611-617. doi: 10.1007/s00239-021-10029-x. Epub 2021 Sep 9. J Mol Evol. 2021. PMID: 34505179
-
Anticodon recognition and discrimination by the alpha-helix cage domain of class I lysyl-tRNA synthetase.Biochemistry. 2007 Oct 2;46(39):11033-8. doi: 10.1021/bi700815a. Epub 2007 Aug 31. Biochemistry. 2007. PMID: 17760422 Free PMC article.
-
Translation termination in pyrrolysine-utilizing archaea.FEBS Lett. 2009 Nov 3;583(21):3455-60. doi: 10.1016/j.febslet.2009.09.044. Epub 2009 Sep 29. FEBS Lett. 2009. PMID: 19796638 Free PMC article.
References
-
- Arnez, J. G., and D. Moras. 1997. Structural and functional considerations of the aminoacylation reaction. Trends Biochem. Sci. 22:211-216. - PubMed
-
- Bult, C. J., O. White, G. J. Olsen, L. Zhou, R. D. Fleischmann, G. G. Sutton, J. A. Blake, L. M. FitzGerald, R. A. Clayton, J. D. Gocayne, A. R. Kerlavage, B. A. Dougherty, J.-F. Tomb, M. D. Adams, C. I. Reich, R. Overbeek, E. F. Kirkness, K. G. Weinstock, J. M. Merrick, A. Glodek, J. L. Scott, N. S. M. Geoghagen, J. F. Weidman, J. L. Fuhrmann, D. Nguyen, T. R. Utterback, J. M. Kelley, J. D. Peterson, P. W. Sadow, M. C. Hanna, M. D. Cotton, K. M. Roberts, M. A. Hurst, B. P. Kaine, M. Borodovsky, H.-P. Klenk, C. M. Fraser, H. O. Smith, C. R. Woese, and J. C. Venter. 1996. Complete genome sequence of the methanogenic archaeon. Methanococcus jannaschii. Science 273:1058-1073. - PubMed
-
- Commans, S., M. Lazard, F. Delort, S. Blanquet, and P. Plateau. 1998. tRNA anticodon recognition and specification within subclass IIb aminoacyl-tRNA synthetases. J. Mol. Biol. 278:801-813. - PubMed
-
- Cusack, S. 1997. Aminoacyl-tRNA synthetases. Curr. Opin. Struct. Biol. 7:881-889. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

