Overcoming drug resistance in HIV-1 chemotherapy: the binding thermodynamics of Amprenavir and TMC-126 to wild-type and drug-resistant mutants of the HIV-1 protease
- PMID: 12142445
- PMCID: PMC2373686
- DOI: 10.1110/ps.0206402
Overcoming drug resistance in HIV-1 chemotherapy: the binding thermodynamics of Amprenavir and TMC-126 to wild-type and drug-resistant mutants of the HIV-1 protease
Abstract
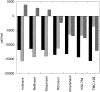
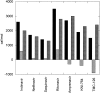
Amprenavir is one of six protease inhibitors presently approved for clinical use in the therapeutic treatment of AIDS. Biochemical and clinical studies have shown that, unlike other inhibitors, Amprenavir is severely affected by the protease mutation I50V, located in the flap region of the enzyme. TMC-126 is a second-generation inhibitor, chemically related to Amprenavir, with a reported extremely low susceptibility to existing resistant mutations including I50V. In this paper, we have studied the thermodynamic and molecular origin of the response of these two inhibitors to the I50V mutation and the double active-site mutation V82F/I84V that affects all existing clinical inhibitors. Amprenavir binds to the wild-type HIV-1 protease with high affinity (5.0 x 10(9) M(-1) or 200 pM) in a process equally favored by enthalpic and entropic contributions. The mutations I50V and V82F/I84V lower the binding affinity of Amprenavir by a factor of 147 and 104, respectively. TMC-126, on the other hand, binds to the wild-type protease with extremely high binding affinity (2.6 x 10(11) M(-1) or 3.9 pM) in a process in which enthalpic contributions overpower entropic contributions by almost a factor of 4. The mutations I50V and V82F/I84V lower the binding affinity of TMC-126 by only a factor of 16 and 11, respectively, indicating that the binding affinity of TMC-126 to the drug-resistant mutants is still higher than the affinity of Amprenavir to the wild-type protease. Analysis of the data for TMC-126 and KNI-764, another second-generation inhibitor, indicates that their low susceptibility to mutations is caused by their ability to compensate for the loss of interactions with the mutated target by a more favorable entropy of binding.
Figures





Similar articles
-
The binding energetics of first- and second-generation HIV-1 protease inhibitors: implications for drug design.Arch Biochem Biophys. 2001 Jun 15;390(2):169-75. doi: 10.1006/abbi.2001.2333. Arch Biochem Biophys. 2001. PMID: 11396919
-
A structural and thermodynamic escape mechanism from a drug resistant mutation of the HIV-1 protease.Proteins. 2004 May 15;55(3):594-602. doi: 10.1002/prot.20069. Proteins. 2004. PMID: 15103623
-
A contribution to the drug resistance mechanism of darunavir, amprenavir, indinavir, and saquinavir complexes with HIV-1 protease due to flap mutation I50V: a systematic MM-PBSA and thermodynamic integration study.J Chem Inf Model. 2013 Aug 26;53(8):2141-53. doi: 10.1021/ci4002102. Epub 2013 Jul 24. J Chem Inf Model. 2013. PMID: 23834142
-
Structural and thermodynamic basis of resistance to HIV-1 protease inhibition: implications for inhibitor design.Curr Drug Targets Infect Disord. 2003 Dec;3(4):311-28. doi: 10.2174/1568005033481051. Curr Drug Targets Infect Disord. 2003. PMID: 14754432 Review.
-
Thermodynamic rules for the design of high affinity HIV-1 protease inhibitors with adaptability to mutations and high selectivity towards unwanted targets.Int J Biochem Cell Biol. 2004 Sep;36(9):1787-99. doi: 10.1016/j.biocel.2004.02.021. Int J Biochem Cell Biol. 2004. PMID: 15183345 Review.
Cited by
-
Thermodynamic and computational analyses reveal the functional roles of the galloyl group of tea catechins in molecular recognition.PLoS One. 2018 Oct 11;13(10):e0204856. doi: 10.1371/journal.pone.0204856. eCollection 2018. PLoS One. 2018. PMID: 30307946 Free PMC article.
-
Resistance mechanism of human immunodeficiency virus type-1 protease to inhibitors: A molecular dynamic approach.Mol Biol Res Commun. 2014 Dec;3(4):253-267. Mol Biol Res Commun. 2014. PMID: 27843989 Free PMC article.
-
Targeting the phosphatidylglycerol lipid: An amphiphilic dendrimer as a promising antibacterial candidate.Sci Adv. 2024 Sep 27;10(39):eadn8117. doi: 10.1126/sciadv.adn8117. Epub 2024 Sep 25. Sci Adv. 2024. PMID: 39321303 Free PMC article.
-
Energy, heat, flavours and aromas of Microbial Biotechnology.Microb Biotechnol. 2008 May;1(3):199-201. doi: 10.1111/j.1751-7915.2008.00036.x. Microb Biotechnol. 2008. PMID: 21261838 Free PMC article. No abstract available.
-
HIV-1 protease molecular dynamics of a wild-type and of the V82F/I84V mutant: possible contributions to drug resistance and a potential new target site for drugs.Protein Sci. 2004 Apr;13(4):1108-23. doi: 10.1110/ps.03468904. Protein Sci. 2004. PMID: 15044738 Free PMC article.
References
-
- Ala, P.J., Huston, E.E., Klabe, R.M., McCabe, D.D., Duke, J.L., Rizzo, C.J., Korant, B.D., DeLoskey, R.D., Lam, P.Y.S., Hodge, C.N., and Chang, C.H. 1997. Molecular basis of HIV-1 protease drug resistance: Structural analysis of mutant proteases complexed with cyclic urea inhibitors. Biochemistry 36 1573–1580. - PubMed
-
- Ala, P.J., Huston, E.E., Klabe, R.M., Jadhav, P.K., Lam, P.Y.S., and Chang, C.-H. 1998. Counteracting HIV-1 protease drug resistance: Structural analysis of mutant proteases complexed with XV638 and SD146, cyclic urea amides with broad specificities. Biochemistry 37 15042–15049. - PubMed
-
- Condra, J.H., Schleif, W.A., Blahy, O.M., Gabryelski, L.J., Graham, D.J., Quintero, J.C., Rhodes, A., Robbins, H.L., Roth, E., Shivaprakash, M., et al. 1995. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature 374 569–571. - PubMed
-
- Freire, E. 2002. Designing drugs against heterogeneous targets. Nat. Biotech. 20 15–16. - PubMed
-
- Fukada, H. and Takahashi, K. 1998. Enthalpy and heat capacity changes for the proton dissociation of various buffer components in 0.1M potassium chloride. Proteins 33 159–166. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

