Thermodynamic environments in proteins: fundamental determinants of fold specificity
- PMID: 12142449
- PMCID: PMC2373679
- DOI: 10.1110/ps.0203202
Thermodynamic environments in proteins: fundamental determinants of fold specificity
Abstract
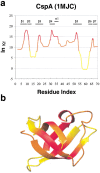
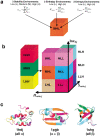
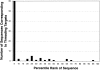
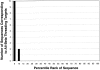
To investigate the relationship between an amino acid sequence and its corresponding protein fold, a database of thermodynamic stability information was assembled as a function of residue type from 81 nonhomologous proteins. This information was obtained using the COREX algorithm, which computes an ensemble-based description of the native state of proteins. Dissection of the COREX stability constant into its fundamental energetic components resulted in 12 thermodynamic environments describing the tertiary architecture of protein folds. Because of the observation that residue types partitioned unequally between these environments, it was hypothesized that thermodynamic environments contained energetic information that connected sequence to fold. To test the significance of this hypothesis, the thermodynamic stability information was incorporated into a three-dimensional-to-one-dimensional scoring matrix, and simple fold recognition experiments were performed in a manner such that information about the fold target was never included in the scoring. For 60 out of 81 fold targets, the correct sequence for the target scored in the top 5% of 3858 decoy sequences, with Z-scores ranging from 1.76 to 12.23. Furthermore, a scoring matrix assembled from the residues of 40 nonhomologous all-alpha proteins was used to thread sequences against 12 nonhomologous all-beta protein targets. In 10 of 12 cases, sequences known to adopt the native all-beta structure scored in the top 5% of 3858 decoy sequences, with Z-scores ranging from 1.99 to 7.94. These results indicate that energetic information encoded by thermodynamic environments represents a fundamental property of proteins that underlies classifications based on secondary structure.
Figures

Similar articles
-
Analysis of the "thermodynamic information content" of a Homo sapiens structural database reveals hierarchical thermodynamic organization.Protein Sci. 2004 Jul;13(7):1787-801. doi: 10.1110/ps.04706204. Protein Sci. 2004. PMID: 15215522 Free PMC article.
-
Thermodynamic propensities of amino acids in the native state ensemble: implications for fold recognition.Protein Sci. 2001 May;10(5):1032-45. doi: 10.1110/ps.01601. Protein Sci. 2001. PMID: 11316884 Free PMC article.
-
Denatured-state energy landscapes of a protein structural database reveal the energetic determinants of a framework model for folding.J Mol Biol. 2008 Sep 19;381(5):1184-201. doi: 10.1016/j.jmb.2008.06.046. Epub 2008 Jun 24. J Mol Biol. 2008. PMID: 18616947 Free PMC article.
-
Novel sequences propel familiar folds.Structure. 2002 Apr;10(4):447-54. doi: 10.1016/s0969-2126(02)00750-5. Structure. 2002. PMID: 11937049 Review.
-
Knowledge-based potential functions in protein design.Curr Opin Struct Biol. 2002 Aug;12(4):447-52. doi: 10.1016/s0959-440x(02)00346-9. Curr Opin Struct Biol. 2002. PMID: 12163066 Review.
Cited by
-
Ensemble-based methods for describing protein dynamics.Curr Opin Pharmacol. 2010 Dec;10(6):760-9. doi: 10.1016/j.coph.2010.09.014. Epub 2010 Oct 19. Curr Opin Pharmacol. 2010. PMID: 20965786 Free PMC article. Review.
-
Hidden dynamic signatures drive substrate selectivity in the disordered phosphoproteome.Proc Natl Acad Sci U S A. 2020 Sep 22;117(38):23606-23616. doi: 10.1073/pnas.1921473117. Epub 2020 Sep 8. Proc Natl Acad Sci U S A. 2020. PMID: 32900925 Free PMC article.
-
Analysis of the "thermodynamic information content" of a Homo sapiens structural database reveals hierarchical thermodynamic organization.Protein Sci. 2004 Jul;13(7):1787-801. doi: 10.1110/ps.04706204. Protein Sci. 2004. PMID: 15215522 Free PMC article.
-
Investigating homology between proteins using energetic profiles.PLoS Comput Biol. 2010 Mar 26;6(3):e1000722. doi: 10.1371/journal.pcbi.1000722. PLoS Comput Biol. 2010. PMID: 20361049 Free PMC article.
-
Energetic profiling of protein folds.Methods Enzymol. 2009;455:299-327. doi: 10.1016/S0076-6879(08)04211-0. Methods Enzymol. 2009. PMID: 19289211 Free PMC article.
References
-
- Anfinsen, C.B. 1973. Principles that govern the folding of protein chains. Science 181 223–230. - PubMed
-
- Baldwin, R.L. and Rose, G.D. 1999. Is protein folding hierarchic? I: Local structure and peptide folding. Trends Biochem. Sci. 24 26–33. - PubMed
-
- Bonneau, R., Tsai, J., Ruczinski, I., and Baker, D. 2001. Functional Inferences from blind ab initio protein structure predictions. J. Struct. Biol. 134 186–190. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

