A large dispersed chromosomal region required for chromosome segregation in sporulating cells of Bacillus subtilis
- PMID: 12145201
- PMCID: PMC126140
- DOI: 10.1093/emboj/cdf393
A large dispersed chromosomal region required for chromosome segregation in sporulating cells of Bacillus subtilis
Abstract
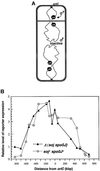
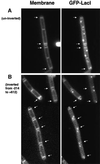
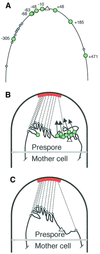
The cis-acting sequences required for chromosome segregation are poorly understood in most organisms, including bacteria. Sporulating cells of Bacillus subtilis undergo an unusual asymmetric cell division during which the origin of DNA replication (oriC) region of the chromosome migrates to an extreme polar position. We have now characterized the sequences required for this migration. We show that the previously characterized soj-spo0J chromosome segregation system is not essential for chromosome movement to the cell pole, so this must be driven by an additional segregation mechanism. Observations on a large set of precisely engineered chromosomal inversions and translocations have identified a polar localization region (PLR), which lies approximately 150-300 kbp to the left of oriC. Surprisingly, oriC itself has no involvement in this chromosome segregation system. Dissection of the PLR showed that it has internal functional redundancy, reminiscent of the large diffuse centromeres of most eukaryotic cells.
Figures



References
-
- Autret S., Nair,R. and Errington,J. (2001) Genetic analysis of the chromosome segregation protein Spo0J of Bacillus subtilis: evidence for separate domains involved in DNA binding and interactions with Soj protein. Mol. Microbiol., 41, 743–755. - PubMed
-
- Bath J., Wu,L.J., Errington,J. and Wang,J.C. (2000) Role of Bacillus subtilis SpoIIIE in DNA transport across the mother cell–prespore division septum. Science, 290, 995–997. - PubMed
-
- Cervin M.A., Spiegelman,G.B., Raether,B., Ohlsen,K., Perego,M. and Hoch,J.A. (1998) A negative regulator linking chromosome segregation to developmental transcription in Bacillus subtilis. Mol. Microbiol., 29, 85–95. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

