Targeting of SWI/SNF chromatin remodelling complexes to estrogen-responsive genes
- PMID: 12145209
- PMCID: PMC126156
- DOI: 10.1093/emboj/cdf412
Targeting of SWI/SNF chromatin remodelling complexes to estrogen-responsive genes
Abstract
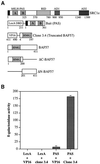
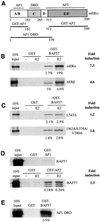
SWI/SNF complexes are ATP-dependent chromatin remodelling enzymes that have been implicated in the regulation of gene expression in yeast and higher eukaryotes. BRG1, a catalytic subunit in the mammalian SWI/SNF complex, is required for transcriptional activation by the estrogen receptor, but the mechanisms by which the complex is recruited to estrogen target genes are unknown. Here, we have identified an interaction between the estrogen receptor and BAF57, a subunit present only in mammalian SWI/SNF complexes, which is stimulated by estrogen and requires both a functional hormone-binding domain and the DNA-binding region of the receptor. We also found an additional interaction between the p160 family of coactivators and BAF57 and demonstrate that the ability of p160 coactivators to potentiate transcription by the estrogen receptor is dependent on BAF57 in transfected cells. Moreover, chromatin immunoprecipitation assays demonstrated that BAF57 is recruited to the estrogen-responsive promoter, pS2, in a ligand-dependent manner. These results suggest that one of the mechanisms for recruiting SWI/SNF complexes to estrogen target genes is by means of BAF57.
Figures






References
-
- Belandia B. and Parker,M.G. (2000) Functional interaction between the p160 coactivator proteins and the transcriptional enhancer factor family of transcription factors. J. Biol. Chem., 275, 30801–30805. - PubMed
-
- Braunstein M., Rose,A.B., Holmes,S.G., Allis,C.D., Broach,J.R., Tisell,L., Fletterick,R.J., Parker,M.G. and Chatterjee,V.K. (1993) Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev., 7, 592–604. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

