Biochemical and genetic characterization of mundticin KS, an antilisterial peptide produced by Enterococcus mundtii NFRI 7393
- PMID: 12147478
- PMCID: PMC124038
- DOI: 10.1128/AEM.68.8.3830-3840.2002
Biochemical and genetic characterization of mundticin KS, an antilisterial peptide produced by Enterococcus mundtii NFRI 7393
Abstract
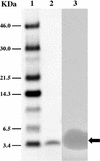
Mundticin KS, a bacteriocin produced by Enterococcus mundtii NFRI 7393 isolated from grass silage in Thailand, is active against closely related lactic acid bacteria and the food-borne pathogen Listeria monocytogenes. In this study, biochemical and genetic characterization of mundticin KS was done. Mundticin KS was purified to homogeneity by ammonium sulfate precipitation, sequential ion-exchange chromatography, and solid-phase extraction. The gene cluster (mun locus) for mundticin KS production was cloned, and DNA sequencing revealed that the mun locus consists of three genes, designated munA, munB, and munC. The munA gene encodes a 58-amino-acid mundticin KS precursor, munB encodes a protein of 674 amino acids involved in translocation and processing of the bacteriocin, and munC encodes a mundticin KS immunity protein of 98 amino acids. Amino acid and nucleotide sequencing revealed the complete, unambiguous primary structure of mundticin KS; mundticin KS comprises a 43-amino-acid peptide with an amino acid sequence similar to that of mundticin ATO6 produced by E. mundtii ATO6. Mundticin KS and mundticin ATO6 are distinguished by the inversion of the last two amino acids at their respective C termini. These two mundticins were expressed in Escherichia coli as recombinant peptides and found to be different in activity against certain Lactobacillus strains, such as Lactobacillus plantarum and Lactobacillus curvatus. Mundticin KS was successfully expressed by transformation with the recombinant plasmid containing the mun locus in heterogeneous hosts such as E. faecium, L. curvatus, and Lactococcus lactis. Based on our results, the mun locus is located on a 50-kb plasmid, pML1, of E. mundtii NFRI 7393.
Figures




References
-
- Bennik, M. H., B. Vanloo, R. Brasseur, L. G. Gorris, and E. J. Smid. 1998. A novel bacteriocin with a YGNGV motif from vegetable-associated Enterococcus mundtii: full characterization and interaction with target organisms. Biochim. Biophys. Acta 1373:47-58. - PubMed
-
- de Vos, W. M., O. P. Kuipers, J. R. van der Meer, and R. J. Siezen. 1995. Maturation pathway of nisin and other lantibiotics: post-translationally modified antimicrobial peptides exported by gram-positive bacteria. Mol. Microbiol. 17:427-437. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

